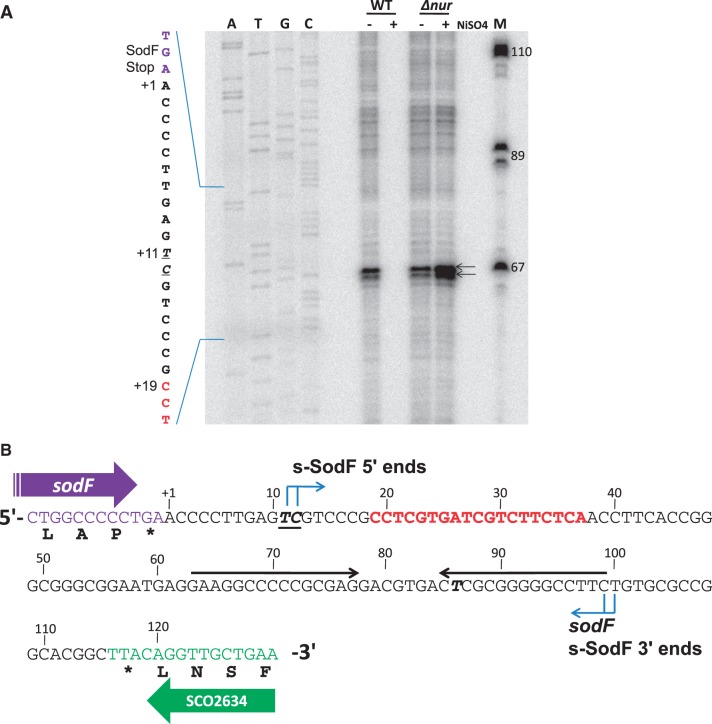
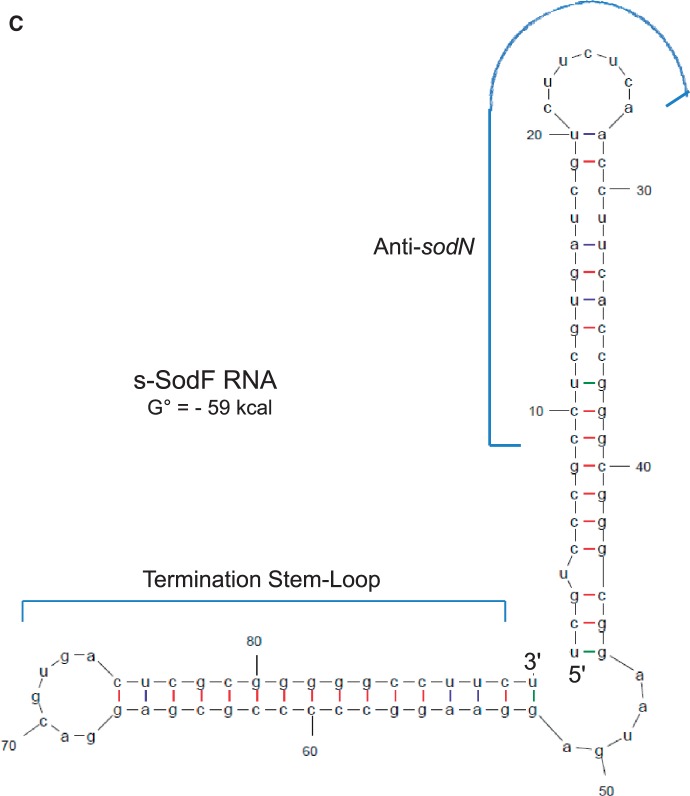
Figure 3.
Determination of 5′-ends of s-SodF RNA. (A) Determination of 5′-ends by high-resolution S1 analysis. S1 mapping of sodF RNAs with a DNA probe 5′-end labeled at +78 nucleotide position relative to the stop codon of the sodF ORF. RNAs were prepared from the wild type (M145) and △nur mutant grown in the presence and absence of 50 μM NiSO4. The position of 5′-ends of the transcripts is underlined. The sequencing ladder was obtained with the sodF oligonucleotide primer (5′-CCT CGC GGG GGC CTT CCT CA-3′) and the template pGEM-sodF346. (B) Sequence information of intergenic region between sodF (SCO2633) and SCO2634. The nucleotide position was numbered relative to the end of the stop (TGA) codon of sodF ORF. The 5′-end position of s-SodF was marked (bold italic underlined) along with 19 nt anti-sodN sequence (red), 15 nt inverted repeats (horizontal arrows), and the possible 3′-end position of sodF and s-SodF RNAs. (C) Predicted secondary structure for s-SodF RNA. The secondary structure of 90 nt long s-SodF RNA (from +11 to +100; as in (B)), as well as stability, were predicted by mfold program (41). The 5′ proximal stem-loop contains the anti-sodN sequence.