Figure 5.
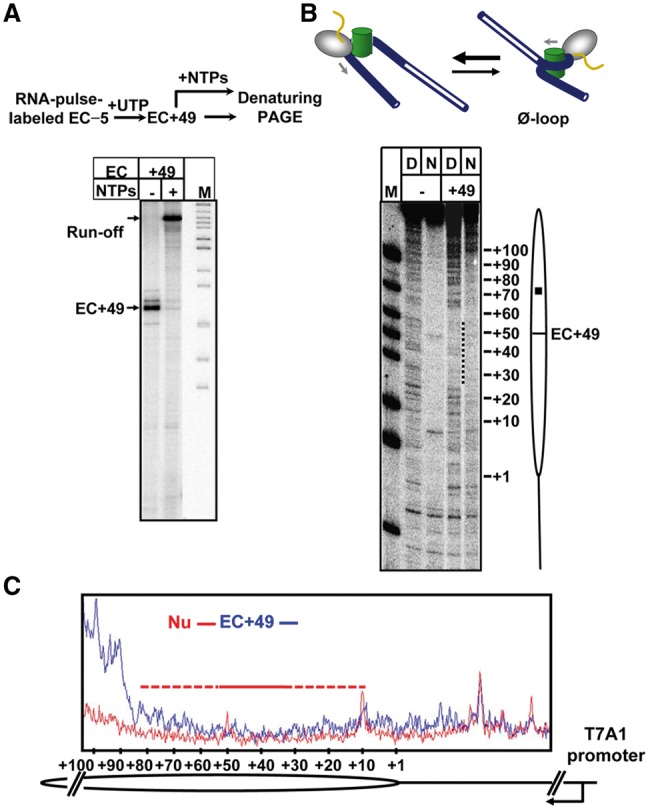
EC+49 formed by T. aquaticus RNAP does not contain the Ø-loop. (A) Analysis of the homogeneity and stability of the EC+49. Top: The experimental approach. Bottom: The EC+49 was formed on the 603 nucleosome and then extended in the presence of all NTPs. Analysis of pulse-labeled RNA by denaturing PAGE. Note that the EC+49 is functional and contains extendable RNA. (B) The structures of the EC+49 formed on histone-free DNA (D) or nucleosomal templates (N) were analysed by DNase I footprinting. The footprints of the EC+49 (dotted line) and the position of the active center of the RNAP are indicated. Analysis of end-labeled DNA by denaturing PAGE. The suggested structure of the EC+49 complex formed on the 603 nucleosome is shown on the top. (C) Quantitative analysis of DNase I footprinting data. Lanes 3 (Nu) and 4 (EC+49 in the 603 nucleosome) in Figure 5B were scanned and the scans were aligned pairwise. Note that formation of the EC+49 results in full DNA uncoiling at the +(1–10) and +(80–150) regions and partial uncoiling at +(10–30) and +(50–80) regions (red-dashed line). The (+30–50) region is strongly protected by RNAP (red solid line). The positions of the nucleosome (oval), nucleosomal dyad (square) and the T7A1 promoter are indicated.
