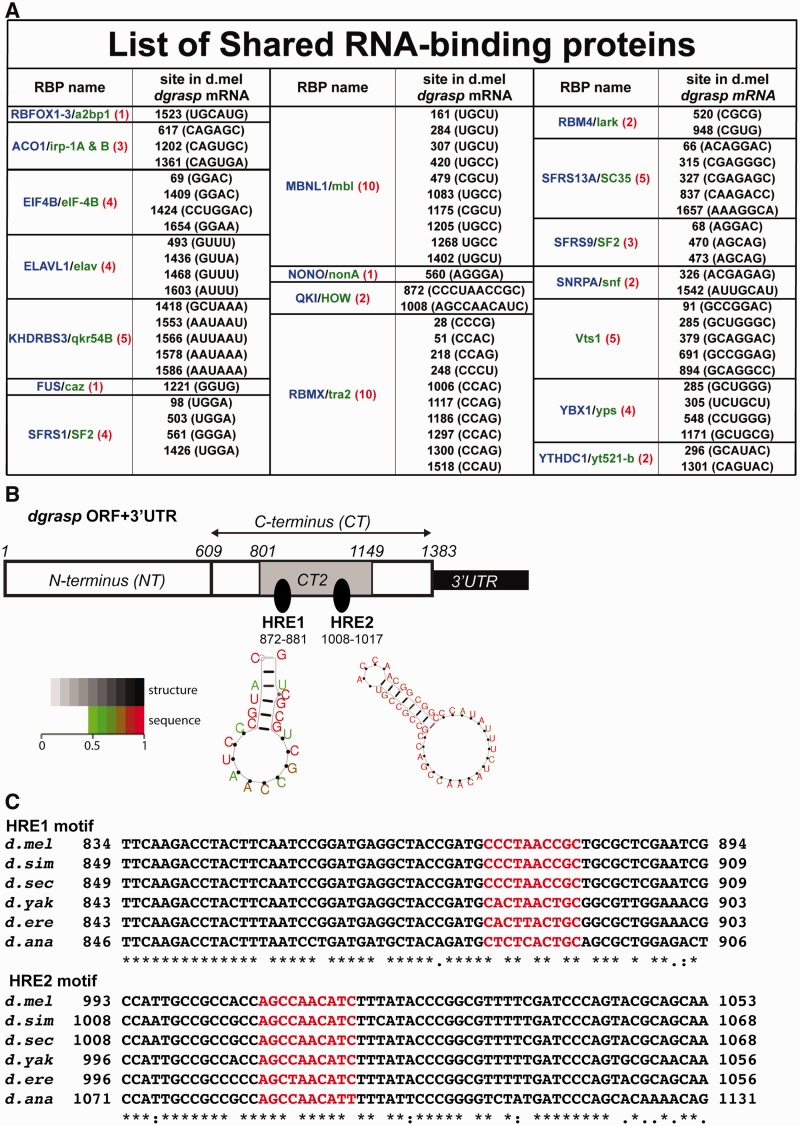
Figure 1.
Computational prediction of HREs in dgrasp mRNA ORF. (A) Outcome of the 18 predicted RBPs from the comparative RBPDB in silico analysis using dgrasp mRNA sequence (ORF and 3′UTR) from D. melanogaster, D. simulans, D. sechellia, D. ananassae, D. erecta and D. yakuba. The vertebrate RBPs are listed in blue whereas the D. melanogaster homologues are in green. The numbers of predicted sites for each RBP, with respect to the translational start site (ATG), are shown in red. (B) Cartoon representation of the two HOW Response Elements (HREs) predicted at the C-terminus of the ORF of D. melanogaster dgrasp at position 872–881 nt (HRE1) and 1008–1017 nt (HRE2) with respect to the translational start site (ATG). The structure prediction of the two HREs using the RNA secondary structure program RNA promo (Segal lab http://genie.weizmann.ac.il/pubs/rnamotifs08/rnamotifs08_predict.html) is presented. The two HRE sites are marked by arrows and are predicted to be in loops. Sequence positions in the predicted structure are colour-coded according to their probability (>0.5 by default) with scale ranging from green (low probability = 0.5) to red (high probability = 1). (C) Alignment of the coding regions of the dgrasp drosophilid orthologues. The two predicted HRE sequences are coloured in red. dgrasp coding sequences of D. melanogaster, D. simulans, D. sechellia, D. ananassae, D. erecta and D. yakuba were used for the ClustalW2-based alignment.