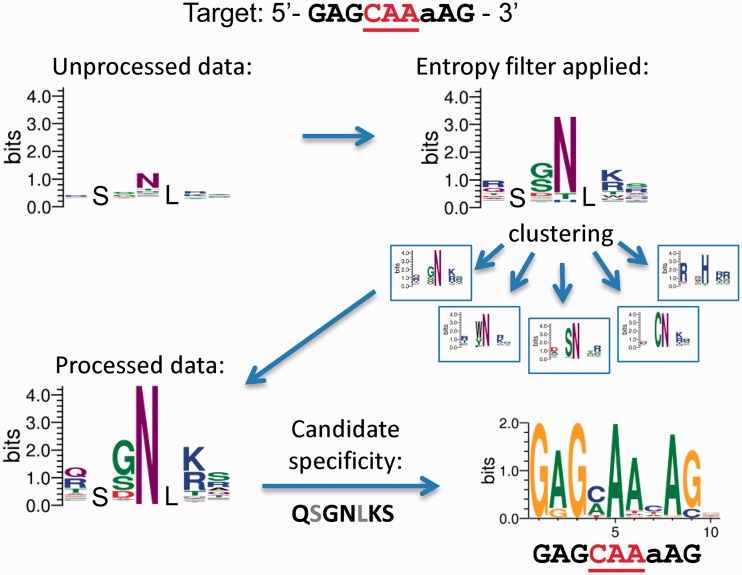
Figure 3.
Overview of the computational pipeline for zinc finger selection analysis. The example details selection of the central finger in a three-fingered array to recognize the 10 bp target 5′-GAGCAA(a/c)AG-3′. The C-terminal and N-terminal constant fingers use the RSANLVR and RSDNLRA helices, which bind GAG and (a/c)AG, respectively. Five amino acid positions (-1, 2, 3, 5 and 6) were randomized with a constant serine at position 1 and leucine at position 4. An unprocessed data sequence logo reflects the raw counts of recovered sequences at five variable positions (top left). Next, after the entropy-based diversity filter is applied, the amino acid enrichment is shown as a sequence logo. Zinc fingers with similar amino acid profiles are clustered and the highly populated cluster used to determine a candidate protein to test specificity (QSGNLKS sequence for the finger 2 positions -1–6) is shown. Finally, the desired binding specificity is confirmed for finger arrays that use a candidate finger that represents the main cluster.