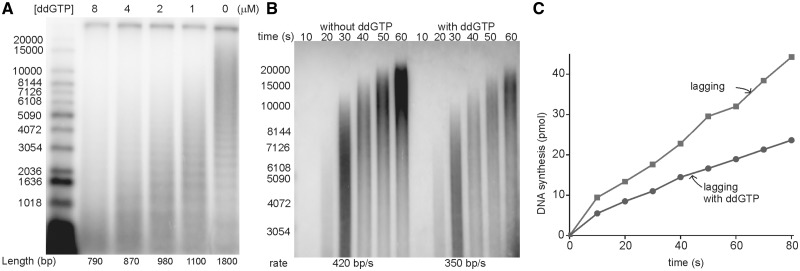
Figure 4.
The availability of primers signals the lagging strand polymerase to cycle. (A) Optimized rolling circle reactions were carried out as described except 120 nM synthetic 15-mer primers substituted for primase and GTP, CTP and UTP. ddGTP was added to the designated concentrations at the same time radiolabeled nucleotide was added. Okazaki fragments incorporating α-[32P] dGTP (5000 cpm/pmol) were monitored by alkaline agarose gel electrophoresis. Lengths were determined with a 20-kb cutoff to exclude leading strand products. (B) In the presence or absence of 4 µM ddGTP, optimized rolling circle reactions were conducted as in (A) except α-[32P] dCTP was added at the same time with ATP and dNTPs. The 3 min elongation step in the presence of non-radioactive dNTPs before the addition of radioactive nucleotide was skipped so that the products would be short enough for accurate length quantification. Leading strand products incorporating α-[32P] dCTP (20 000 cpm/pmol) were monitored by alkaline agarose gel electrophoresis. The products of 20, 30 and 40 s were used to calculate the rate of leading strand synthesis. (C) The amount of lagging strand synthesis in the presence of 1 µM ddGTP was quantified.