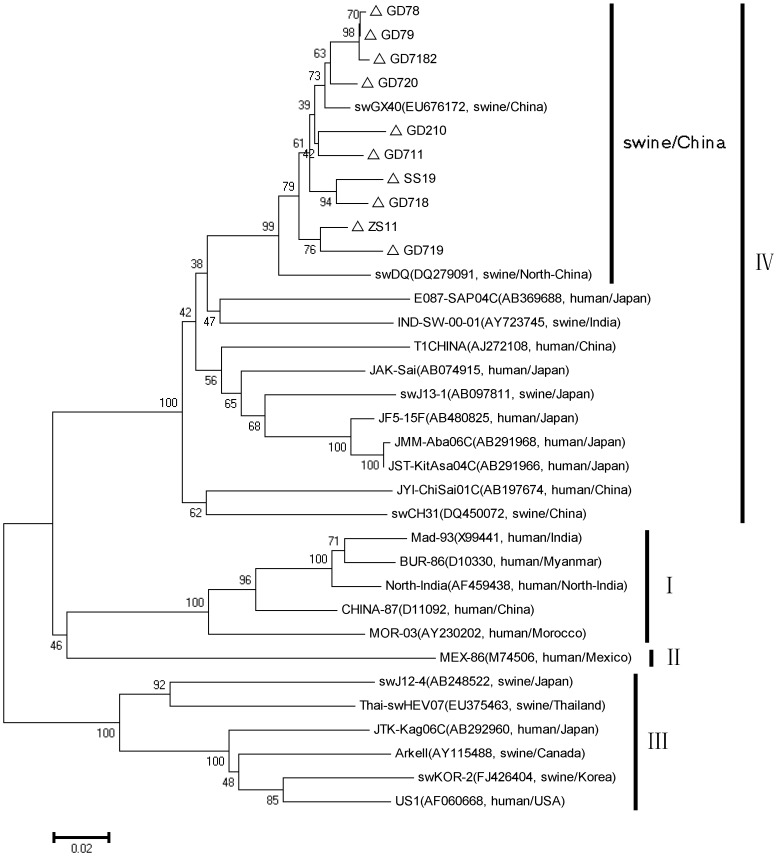
Figure 1. A phylogenetic tree based on the full nucleotide sequence of HEV.
Total RNA positive bile samples are 48, from 9 different districts in Guangdong. We have chosen at least one sample from each district by random sampling. The amplification products of ORF2 (509 nucleotides, primer sequences were HEV-INF and HEV-INR as described above) from 10 positive bile samples were sequenced and compared. The nucleotide sequence identity among the 10 swine HEV isolates obtained from pigs from different farms in three years ranged between 94.3 and 99.8%. Phylogenetic trees were constructed by the neighbor-joining method based on the partial nucleotide sequence of the ORF2 region (509 nucleotides). The bootstrap values (expressed as percentages) were determined on 1000 re-samplings of the data sets. △were the isolates in this study.