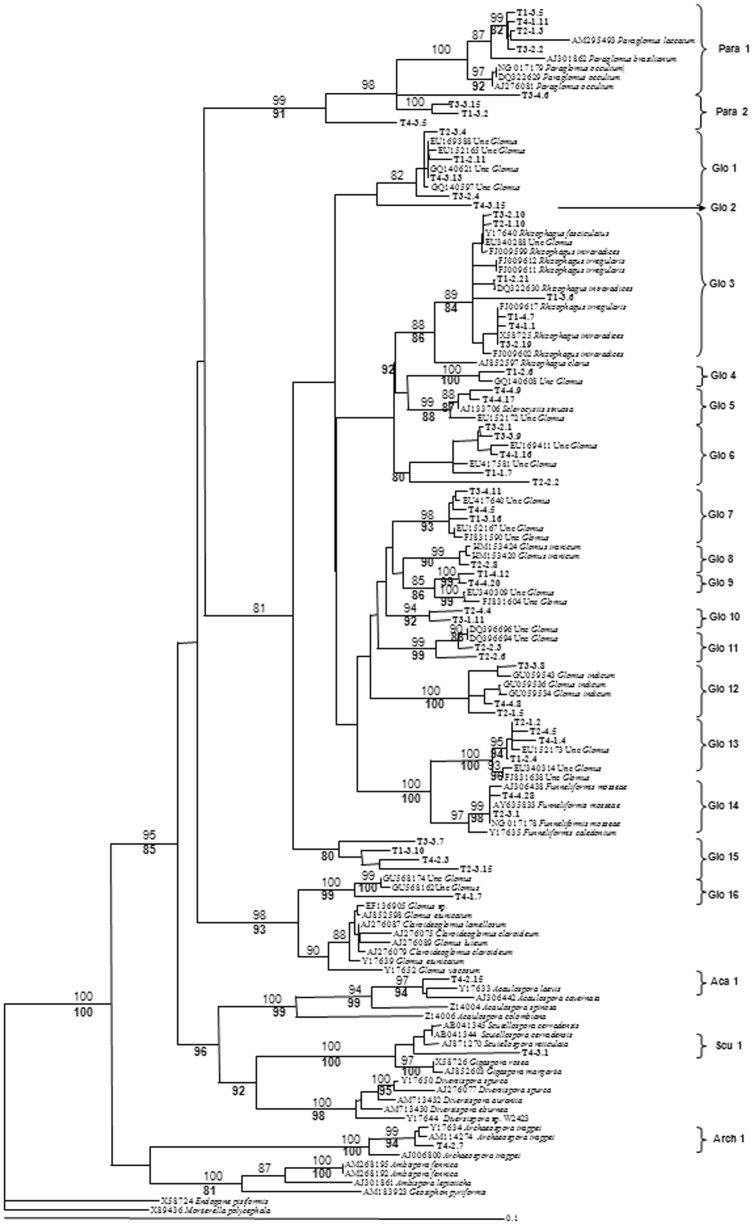
Figure 1. Phylogenetic tree of AMF sequences isolated from the Prunus persica roots under different treatments (T1: Combination of organic and inorganic fertilization and integrated pest management; T2: Inorganic fertilization and integrated pest management; T3: Inorganic fertilization and chemical pest control; T4: Combination of organic and inorganic fertilization and chemical pest control), reference sequences corresponding to the closest matches from GeneBank as well as sequences from cultured AMF taxa including representatives of the major taxonomical groups.
Numbers above branches indicate the bootstrap values determined for Neighbour-Joining (NJ) analysis; bold numbers below branches indicate the bootstrap values of the maximum likelihood analysis. Sequences are labelled with the number of treatment from which they were obtained (T1, T2, T3, T4) and the clone identity number, Group identifiers (for example Glo 1) are AM fungal sequences types found in our study. Since identical sequences were detected, the clones producing the same sequence for each treatment were represented once in the alignment for clarity (Table S1 in File S1 material show a detailed description of the total number of clones of each AMF phylotype that were recovered from each treatment).