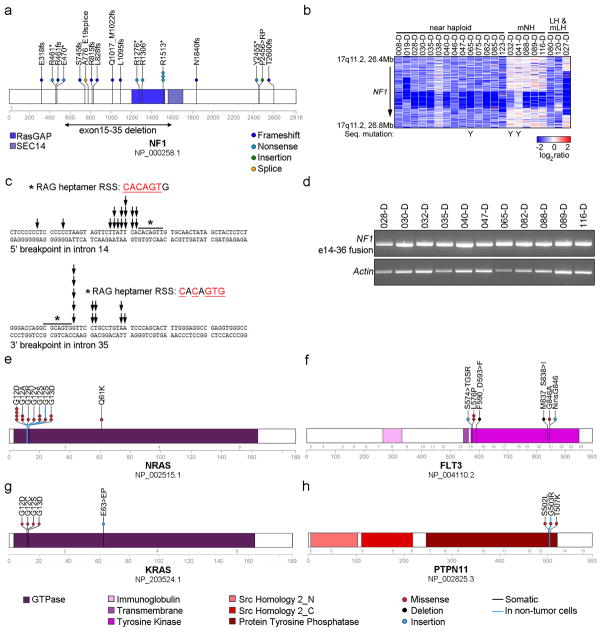
Figure 3. Recurrent alterations of Ras- and RTK signaling in near haploid ALL.
a, Protein domain plot of NF1 depicting identified alterations. The recurrent exon 15–35 deletion in NF1 is indicated by a double-headed arrow. b, SNP 6.0 microarray heatmap showing deletions in NF1. Blue indicates DNA loss. Cases with simultaneous NF1 deletion and sequence mutation are indicated by a Y. Masked hypodiploid cases here include all cases harboring a doubled clone constituting at least 30%. mNH, masked near haploid; LH, low hypodiploid; mLH, masked low hypodiploid. c, The genomic breakpoints for the NF1 deletions cluster in two regions. The individual breakpoints are indicated by arrows. The 5′-break (upper panel) is most frequently present in intron 14, and the 3′-break (lower panel) in intron 35. Immediately internal to the 5′- and 3′-breakpoints are putative, partially conserved RAG heptamer recombination signal sequences (RSS) present, and an insertion of a variable number of non-consensus nucleotides in between the two breakpoints were seen for all these cases. d, RT-PCR on cases harboring an NF1 exon 15–35 deletion, using forward and reverse primers complementary to regions in NF1 exons 14 and 36, respectively, rendering a 648 bp fragment. e–h, Protein domain- and alteration plots for other Ras- and RTK signaling related genes recurrently targeted by sequence mutations in hypodiploid ALL, including NRAS, KRAS, FLT3 and PTPN11 (schematic of MAPK1 can be found in Supplementary Fig. 4). Blue line indicates alterations present also in non-tumor cells.