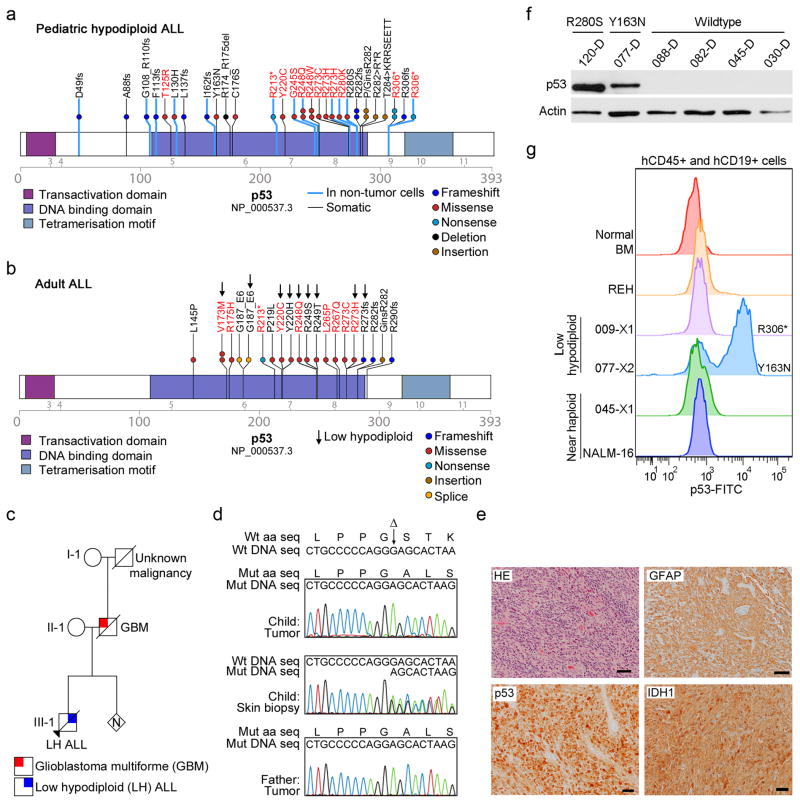
Figure 4. Frequent mutations in TP53 in low hypodiploid ALL.
a–b, Protein domain plots of p53 with alterations identified in (a) pediatric hypodiploid ALL and (b) adult ALL. Known LFS alterations are indicated in red. Alterations present in non-tumor cells are indicated by blue lines. Arrows indicate alterations identified in adult low hypodiploid cases in b. c, Pedigree of a family with an inherited TP53 mutation. N, number of siblings. d, Electropherograms showing the inherited TP53 g.13886delG mutation (resulting in p.G302fs) in the proband (child; leukemic bone marrow (top) and normal skin biopsy(middle)) and father (lower panel). Wild-type (Wt) DNA and amino acid (aa) sequences (seq) are depicted at the top of the panel. Δ indicates the deleted nucleotide. e, Histologic examination of tumor from the proband’s father indicated a diagnosis of glioblastoma. Poorly differentiated glial cells are admixed with sparse cells showing an astrocytic phenotype. Many neoplastic cells are GFAP-immunopositive, and all express the mutant TP53 and IDH1 (p.Arg132His) gene products. Scale bar corresponds to 50 microns. HE, hematoxylin and eosin. f, Immunoblot of p53 on primary hypodiploid ALL samples harboring either wild-type TP53 or a TP53 missense mutation as indicated. g, Flow cytometric analysis of spleen samples from xenografted mice and NALM-16 detecting p53 levels. Only cells positive for human CD45 and CD19 were analyzed. Known nonsense and missense alterations in p53 are indicated. BM, bone marrow.