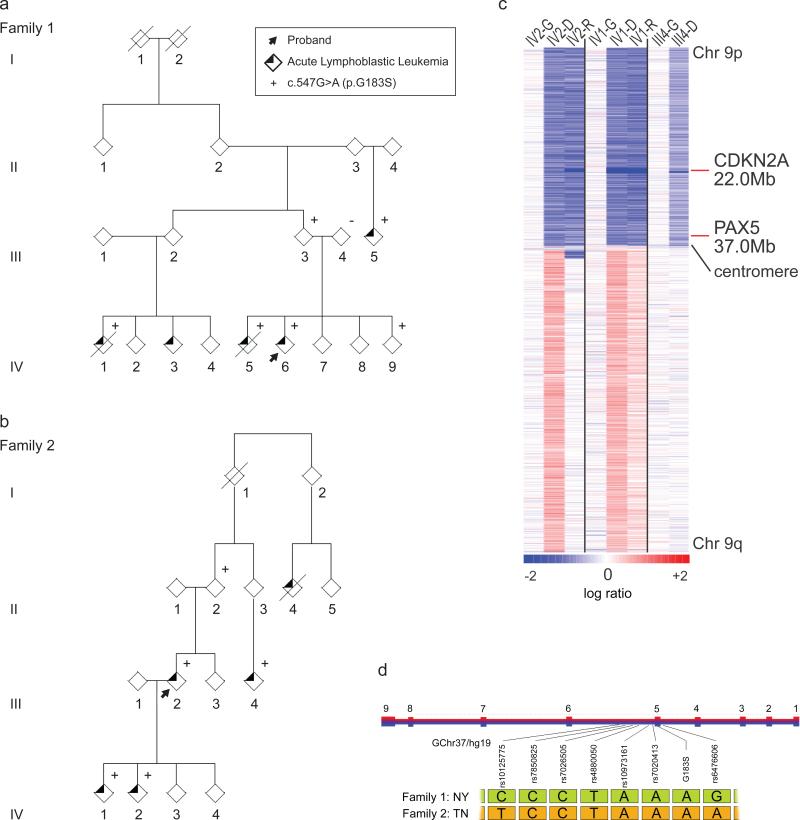
Figure 1. Familial Pre-B cell ALL associated with i(9)(q10) and dic(9;v) in two families harboring a novel, recurrent germline p.Gly183Ser variant.
a. Family 1 of Puerto Rican ancestry. The proband is denoted by an arrow. Exome sequencing was undertaken in germline DNA from all available affected (IV1, IV5, IV6, III5) and unaffected (IV9, III3, III4) individuals as well as the diagnostic leukemic sample from IV6. p.Gly183Ser variant status denoted by (+/−). b. Family 2 of African-American ancestry. The proband is denoted by an arrow. Exome sequencing was undertaken in diagnostic, remission and relapse leukemic samples from individuals III4, IV1, and IV2. p.Gly183Ser variant status denoted by (+/−). c. Chromosome 9 copy number heat map for SNP6.0 microarray data of germline and tumor samples from three members of Family 2. These data demonstrate the common feature of loss of 9p in the tumor specimens. Note the focal dark blue band denoting homozygous loss of CDKN2A/B in all samples. Blue indicates deletions and red indicates gains. G, germline; D, diagnosis; R, relapse sample. d. The haplotype flanking the p.Gly183Ser mutation. A five SNP haplotype rs7850825 to rs7020413 (Chr9:36.997-37.002 Mb) proximal to the mutation was concordant in both family 1 and family 2. However, the distal end flanking the mutation rs6476606 was discordant.