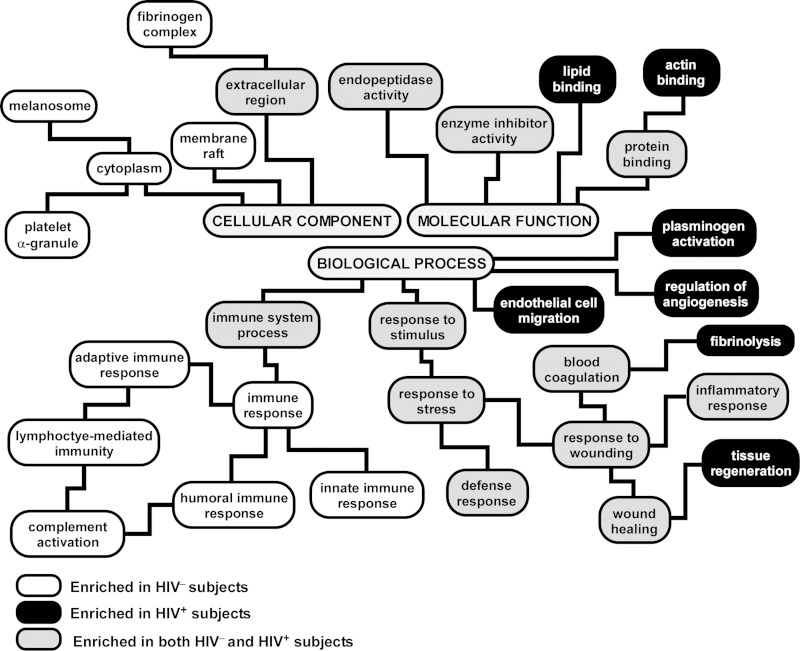
Fig. 3.
Functional categorization of differentially expressed proteins in BALF of HIV+ vs. HIV− subjects based the Gene Ontology (GO) annotations. The hierarchical structure of GO is displayed, with branches showing representative enriched functional pathways (adjusted P value <0.05). A complete list is provided in Supplemental Tables 2 and 3. Note that many processes were common to HIV+ and HIV− individuals (gray boxes), consistent with previous reports that BALF is enriched proteins mapping to distinct functional groups such as endopeptidase activity, defense response, and inflammatory response. However, several immune-related pathways were overrepresented only in HIV− subjects (white boxes), implying depletion of their protein members in HIV+ patients, whereas some processes, such as endothelial cell migration and angiogenesis, were selectively enriched in HIV+ individuals (black boxes).