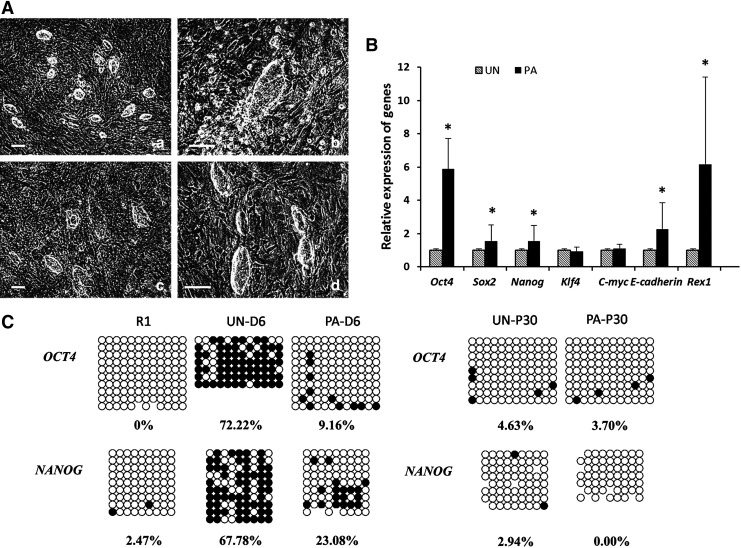
FIG. 3.
Comparison of unpassaged and continuously passaged cells during early reprogramming. (A) Morphology of unpassaged and continuously passaged cells at day 3 after picking clones. a and b, Unpassaged cells; c and d, passaged cells. (B) Expression levels of Oct4, Sox2, Nanog, Klf4, C-myc, Rex1, and E-cadherin genes in unpassaged (UN group) and continuous passaged cells (PA group) relative to GAPDH (a loading control) as assessed by real-time PCR. The values from unpassaged cells were set to 1. The error bar indicates standard deviation (SD). (C) Methylation analysis of OCT4 and NANOG promoter regions in iPSCs. Genomic DNA from R1ES cells, unpassaged cells, and continuously passaged cells after culturing for 6 days, and unpassaged and continuously passaged cells after culturing for passage 30 days. Open and filled circles indicate unmethylated and methylated CpG dinucleotides, respectively. Scale bar, 100 μm.