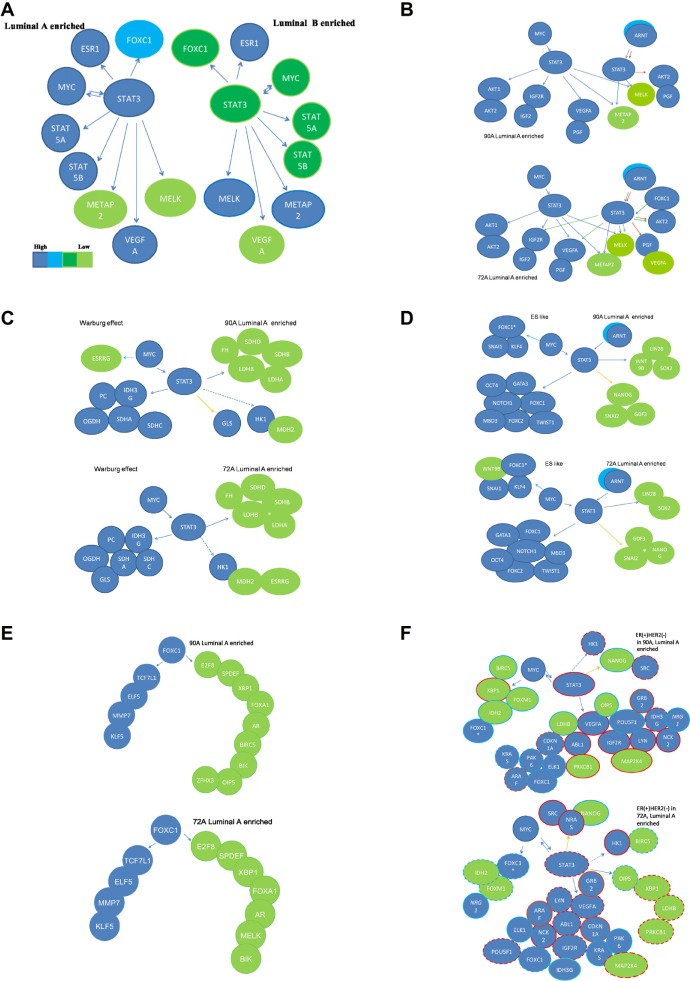
Figure 3.
Functional prediction on roles of STAT3 gene partners in STAT3 subnetworks of ER(+) IDCs.
6 functional subnetworks in 90 A and 72 A cohorts are generated via (1) Predicted networks derived from overlapping genes in Venn diagrams of the STAT3 subnetworks in ER(+) breast cancer gene expression profiles from a 90 A cohort (A); (2) 4 feature functionalities (cell proliferation, sustained angiogenesis, the Warburg effect and ES-like phenotype) of the major STAT3 target genes in 2 STAT3 subnetworks are either commonly co-regulated by MYC (C) and/or differentially co-regulated by FOXC1 and/or ARNT (B and D) in the ER(+) IDCs. A subset of genes, which are predicted to be prognostic factors, is potentially regulated by multiple combined routes of MYC and STAT3, ARNT/HIF1α and STAT3, ARNT/HIF2α and STAT3 or STAT3 for ES-like phenotype (D). E stands for the FOXC1 subnetwork to be a part of activities in cell proliferation. The summary for prognostic features of the STAT3 subnetworks in 90 A cohort and 72 A cohort are shown in F. Solid/dashed lines stand for the specific pathway identified as significant/insignificant in gene expression relationship between a TF and a target gene. Each arrow points toward its downstream target. The combined routes toward the same target gene are labeled with the same color. Relative mRNA expression levels are shown in a color scale (A). Poor prognostic factors are marked by the red rings. Good prognostic factors are marked by the light blue rings. If a probe is not significant shown by Kaplan-Meier survival analysis, it is marked with dotted ring.