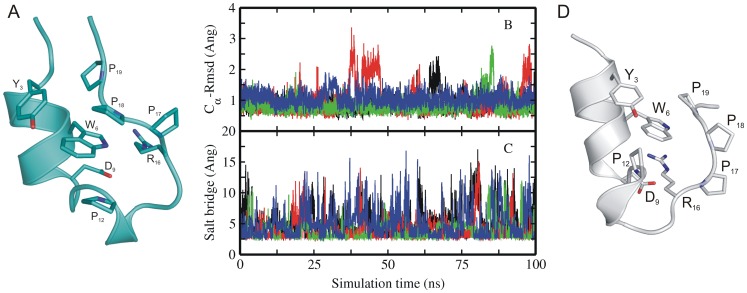
Figure 1. Experimental structure and MD simulations of native Trp-cage miniprotein.
(A) Cartoon representation of the folded experimental structure of Trp-cage mini protein [1]. A subset of side chains important for folding are labeled (stick representation). (B) Root-mean –square deviation (RMSD) of Cα–atoms with respect to native structure and (C) distance between Asp9-Arg16 (salt bridge) of sampled Trp-cage conformations in explicit solvent starting from the native structure (1st entry of pdb 1L2Y) versus simulation time. Simulations were performed with four different force fields encoded as different line colors (black:ff03, red:ff99SB, green:ff99SB_ILDN,blue:ff99SB_NMR). (D) Partially unfolded Trp-cage structures sampled during simulation starting from native state (with the C-terminal PolyProII motive transiently dissociated from the central Trp6 residue).