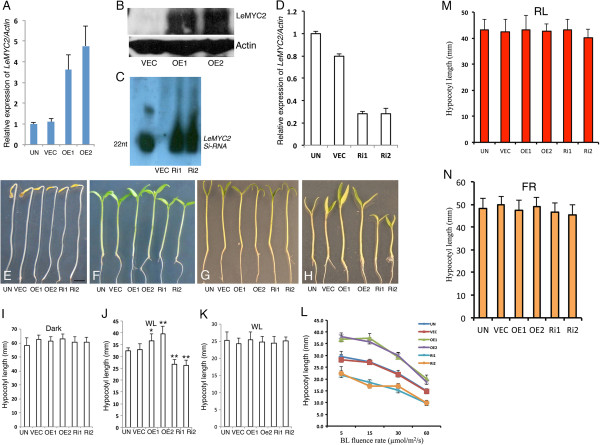
Figure 1.
LeMYC2 mediated regulation of hypocotyl elongation during early tomato seedling development. A, Real time PCR analysis for LeMYC2 transcript levels in 6-day-old seedlings of various LeMYC2 overexpresser transgenic lines grown in BL. LeActin used as control. Error bars represent SD. Number of independent experiments with similar results is (n ≥ 3). B, Western blot analysis of control (VEC) and LeMYC2 overexpresser transgenic lines (OE1 and OE2). The gel blot was probed with anti-MYC2 antibodies. Anti-ACTIN antibody was used to probe ACTIN immunoblot. C, Detection of gene-specific 22 nucleotide siRNAs by Northern blot analysis in RNAi transgenic plants targeting LeMYC2. LeMYC2 was used as probe for the detection of siRNAs. D, Real time PCR analysis for LeMYC2 transcript levels in 6-day-old seedlings of various LeMYC2 RNAitransgenic lines grown in BL. For experimental detail, see legend to A. E to H, Visible phenotypes of 6-day-old seedlings grown in darkness, white light (WL: 5 μmol/m2/s), white light (WL: 30 μmol/m2/s) and blue light (BL: 30 μmol/m2/s), respectively. In each panel, from left to right seedlings of UN (Untransformed), VEC (vector control), OE1 (LeMYC2OE1), OE2 (LeMYC2OE2),Ri1 (LeMYC2RNAi1) and Ri2 (LeMYC2RNAi2) lines are shown. Scale bar, 1 cm. I to L, Quantification of hypocotyl length of 6-day-old seedlings as shown in A to D, respectively. About 15 seedlings of each line were used for the measurement of hypocotyl length. Error bars indicate standard deviation (SD, n = 6). Asterisks in Figure F indicate that OE1, OE2, Ri1 and Ri2 are significantly different from the vector control (*P < 0.05 and **P <0.01, Student’s t test). The experiment was repeated for 4 times. M and N, Quantification of hypocotyl length of 6-day-old seedlings grown in RL (30 μmol/m2/s) and FR (20 μmol/m2/s), respectively. For experimental detail, see legend E to H.