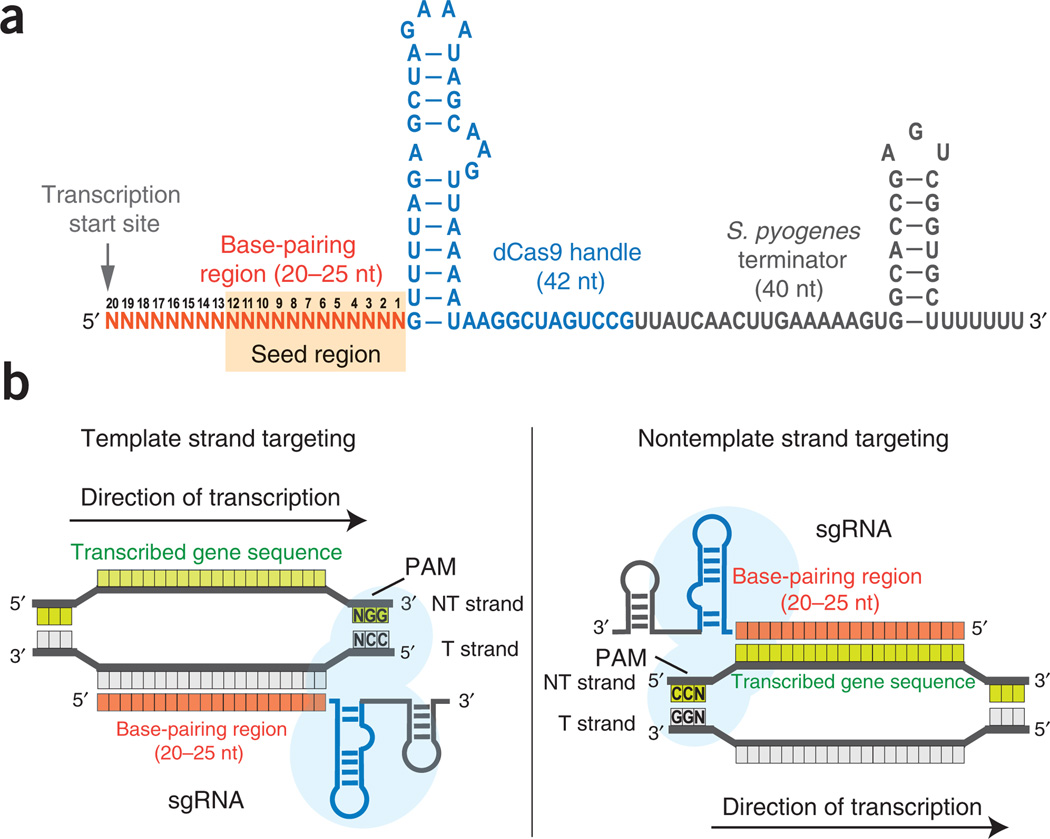
Figure 3.
Design of the sgRNAs. (a) The sgRNA is a chimera and consists of three regions: a 20–25-nt-long base-pairing region for specific DNA binding, a 42-nt-long dCas9 handle hairpin for Cas9 protein binding and a 40-nt-long transcription terminator hairpin derived from S. pyogenes. Transcription of sgRNAs should start precisely at its 5′ end. The 12-nt seed region is shaded in orange. (b) The schemes for designing sgRNAs to target the template (T) or nontemplate (NT) DNA strands. When targeting the template DNA strand, the base-pairing region of the sgRNA has the same sequence identity as the transcribed sequence. When targeting the nontemplate DNA strand, the base-pairing region of the sgRNA is the reverse-complement of the transcribed sequence.