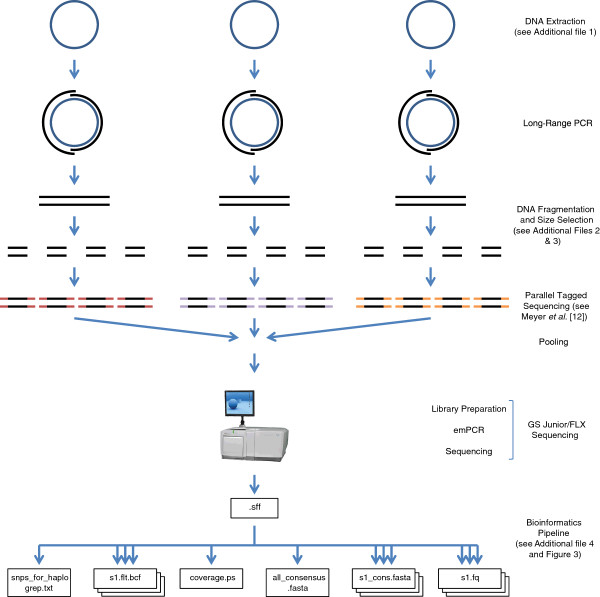
Figure 1.
Overview of the A to Z method for high-throughput DNA sequencing of complete human mitochondrial genomes. DNA is collected using cheek swabs and then extracted using a phenol–chloroform method. Long-range PCR is used to amplify each mitochondrial genome in two overlapping amplicons. The two amplicons from each genome are then pooled and fragmented using NEBNext dsDNA Fragmentase. Barcoding of the fragments is then achieved using Parallel Tagged Sequencing (PTS) [12]. Barcoded fragments are then pooled for library preparation, emulsion PCR (emPCR) and pyrosequencing on the 454 GS FLX platform. Using a number of bioinformatics tools, the resulting sequence data are de-multiplexed and barcodes and primers are removed. Reference-based mapping (to a circular reference) is carried out, followed by the output of coverage plots, consensus sequences and SNP calling for each individual.