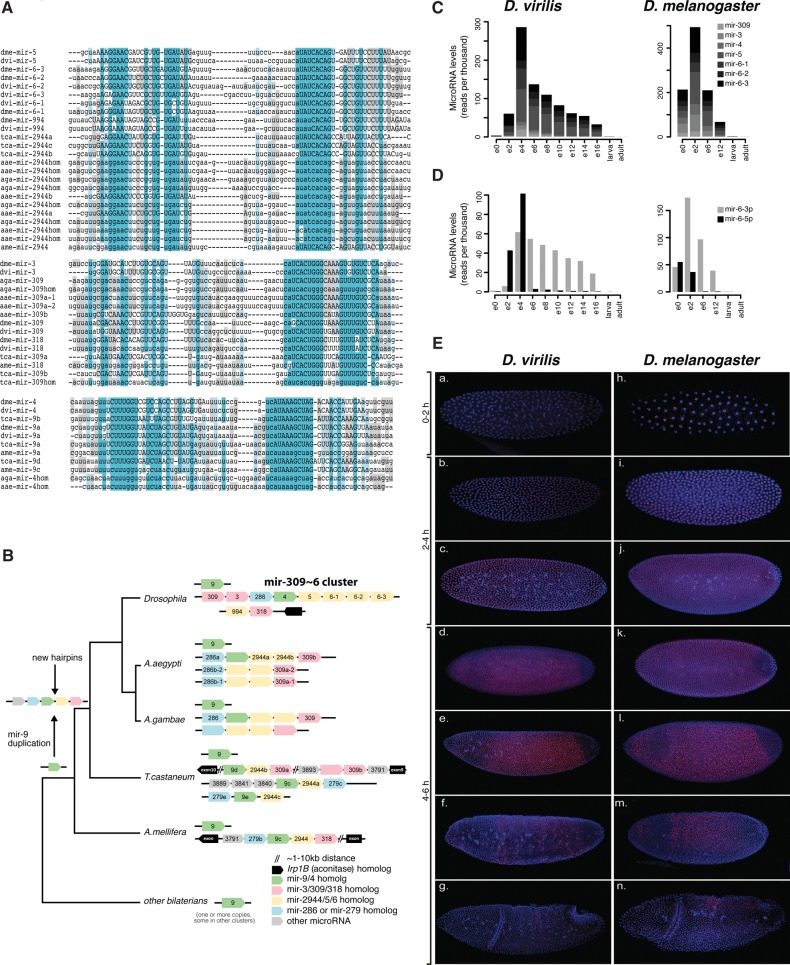
FIGURE 5.
Expression and evolution of the mir-309∼6 cluster. (A) MicroRNA sequence alignments for mir-309/3/318, mir-5/6/2944, and mir-9/mir-4 homologs in D. melanogaster, D. virilis, Anopheles gambiae, Aaedes aegypti, Tribolium castaneum, and A. mellifera. Known mature sequences are shown in uppercase, and conserved sites are highlighted. (hom) Denotes homologs that are discovered in our study but not annotated in miRBase. (B) Evolutionary history of the mir-309∼6 cluster. The organization and content of microRNA clusters containing homologs of members of the mir-309∼6 in Drosophilids are shown for D. melanogaster, D. virilis, A. gambiae, A. aegypti, T. castaneum, and A. mellifera. Black lines represent different chromosomes/scaffolds. MicroRNAs annotated in miRBase are labeled, and homologs are color-coded but blank. Note that some species have one or more additional paralogs of mir-9, which are not shown, for simplicity. (C) Expression levels of mature mir-309∼6 cluster products throughout the development of D. virilis and D. melanogaster. (D) Expression levels of the 5′ and 3′ arms of D.virilis and D. melanogaster mir-6 paralogs throughout the development of the two species. (E) Expression of mir-309∼6 primary transcript in embryos from D. virilis and D. melanogaster. All embryos are oriented with ventral to the top and anterior to the right. The mir-6 primary transcript is expressed ubiquitously in precellular embryos of D. virilis [(a) 0–2 h; (b,c) 2–4 h] and D. melanogaster (h–j), except for the pole bud cells. In stage 5 embryos of both species (d,e,k,l,), the expression is repressed first from the posterior, and next in the anterior region. During the next stages (f,g,m,n), repression continues, until a final dorsal stripe disappears by the time of gastrulation.