Figure 6.
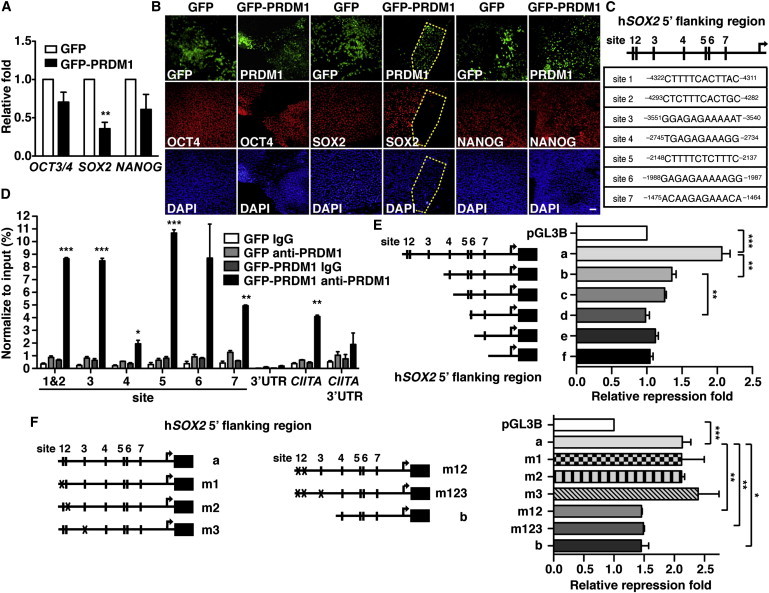
Enforced PRDM1 Expression Represses SOX2
(A) qRT-PCR shows the relative mRNA levels of OCT3/4, SOX2, and NANOG in H9 cells 7 days after transduction with GFP-PRDM1- or GFP-expressing lentiviral vectors.
(B) Immunofluorescence staining of OCT4, SOX2, or NANOG in H9 cells expressing GFP-PRDM1 or GFP for 7 days. Scale bar, 100 μm.
(C) Schematic representation of seven potential PRDM1-binding sites on the SOX2 5′ flanking region. The transcriptional start site (+1) is indicated.
(D) ChIP showed the binding of PRDM1 in the SOX2 5′ flanking region in NCCIT EC cells expressing GFP-PRDM1. NCCIT EC cells were transduced with either GFP-PRDM1 or GFP producing lentiviral vectors. Two days later, ChIP analysis was performed. Immunoglobulin G (IgG) was used as isotype control antibody. The 3′ UTR of SOX2 and CIITA gene loci and CIITA promoter III were used as the negative control and positive loci for PRDM1 binding, respectively. Results (mean ± SEM) are triplicate data for one representative experiment of three independent experiments.
(E and F) The luciferase reporter assay showed that multiple PRDM1-binding sites on the SOX2 5′ flanking region are involved in PRDM1-mediated suppression. Relative luciferase activity was measured using lysates from NCCIT EC cells transfected with PRDM1 expression or control vector, equal molar of the indicated luciferase reporters driven by various lengths of SOX2 5′ flanking region (E) or SOX2 5′ flanking region containing various PRDM1-binding-site mutants (m) (F) together with thymidine kinase promoter-Renilla luciferase reporter plasmid (RL-tk). Luciferase activity assay was analyzed 48 hr later. Results in (A), (E), and (F) are the mean ± SEM of three independent experiments.
See also Figure S3.
