Figure 7.
The PRDM1/SOX2 Regulatory Axis Controls Germ Cell versus Neural Cell Fate in BMP4- and WNT3A-Treated hESCs
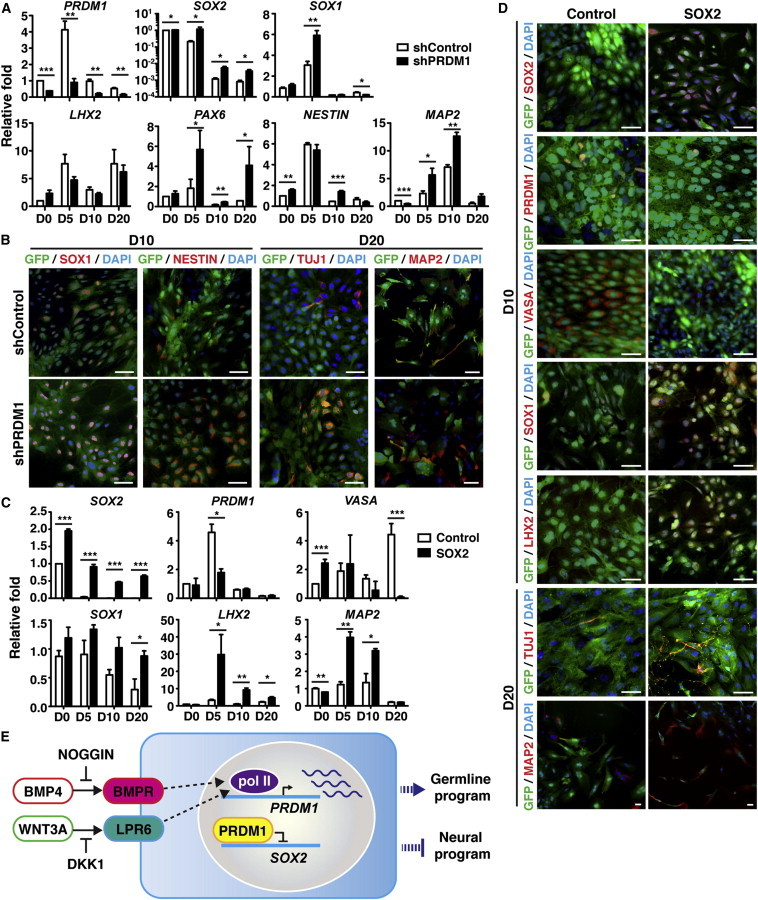
(A) shControl or shPRDM1 lentiviral vectors were transduced into BMP4- and WNT3A-treated H9 cells. Alteration of mRNA expression was quantified by qRT-PCR. Relative values were calculated by comparing the mRNA levels of the indicated genes in PRDM1-KD cells at the indicated days with shControl-transduced cells on day 0.
(B) Immunofluorescence staining indicated that shPRDM1-transduced, but not shControl-transduced, cells showed enhanced SOX1, NESTIN, TUJ1, and MAP2 at the indicated days after BMP4 and WNT3A treatment. Scale bar, 30 μm.
(C) Sorted GFP+ H9 cells that were expressing control GFP alone or SOX2 and GFP simultaneously were treated with BMP4 and WNT3A, followed by qRT-PCR analysis to detect the indicated genes at various days. The fold change was plotted relative to the values obtained from the control vector-transduced cells on day 0.
(D) Immunofluorescence staining showed the expression of exogenous SOX2, reduced expression of PRDM1 and VASA, and increased expression of SOX1, LHX2, TUJ1, and MAP2 as compared with control GFP+ cells at the indicated days after BMP4 and WNT3A treatment. Scale bar, 30 μm. Results in (A) and (C) are the mean ± SEM of three independent experiments.
(E) Working model of the function of PRDM1 in germline differentiation by hESCs. BMP4- and WNT3A-mediated upregulation of PRDM1 transcriptionally suppresses SOX2, thereby inducing the activation of germline fate and preventing the neural program. Dashed lines indicate that the detailed signaling pathways leading to the induction of PRDM1 have not been characterized.
See also Figures S4 and S5.
