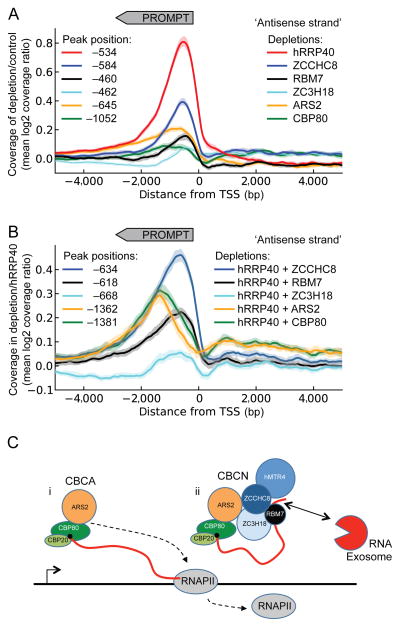
Figure 7.
CBCA depletion displays a genome-wide termination defect of PROMPT transcription (a) Coverage profiles of mean log2(ratio) between indicated factor depletion (color coded) and control (eGFP siRNA) sample plotted on the antisense strand ± 5 kb relative to the annotated gene TSSs. Peak positions of each curve smoothened over 501-bp sliding windows are noted in the left side of the diagram. The direction of PROMPT transcription is indicated by grey arrow-box on top. Faded areas represent 95% confidence intervals for each curve (bootstrap analysis with 1000 resampling replicates). (b) Coverage profiles plotted and presented as in a, but with indicated co-depletions relative to the hRRP40-depleted sample. Dotted lines indicate peak positions of each curve. (c) Model for assembly and utility of CBCN in human nuclear RNA metabolism. i) The CBCA complex associates with the nascent RNA cap and impacts TSS-proximal transcript termination. ii) CBCN assembles on the RNA-CBC platform of these transcripts and recruits the RNA exosome. See text for details.