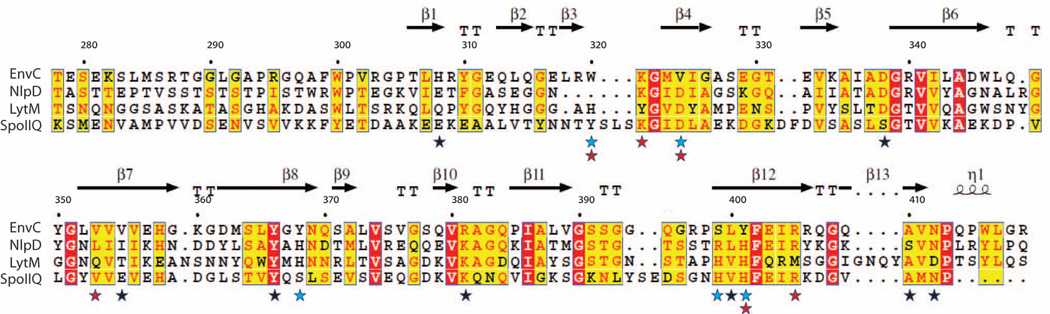
Figure 2. LytEnvC secondary structure map and sequence alignment with other LytM domains.
The secondary structure elements found in theLytEnvC structure are indicated above the sequence alignment. Residue numbering is for EnvC.LytEnvC residues important for function are highlighted with a red star.LytEnvC residues selected for mutagenesis but resulting in unstable mutant proteins are highlighted with a black star. Residues required forSaLytM catalysis are indicated by a blue star. Residues that are conserved or similar in at least three proteins in the alignment are boxed in yellow. Positions that are absolutely conserved are boxed in red. β, β-strand; T, turn