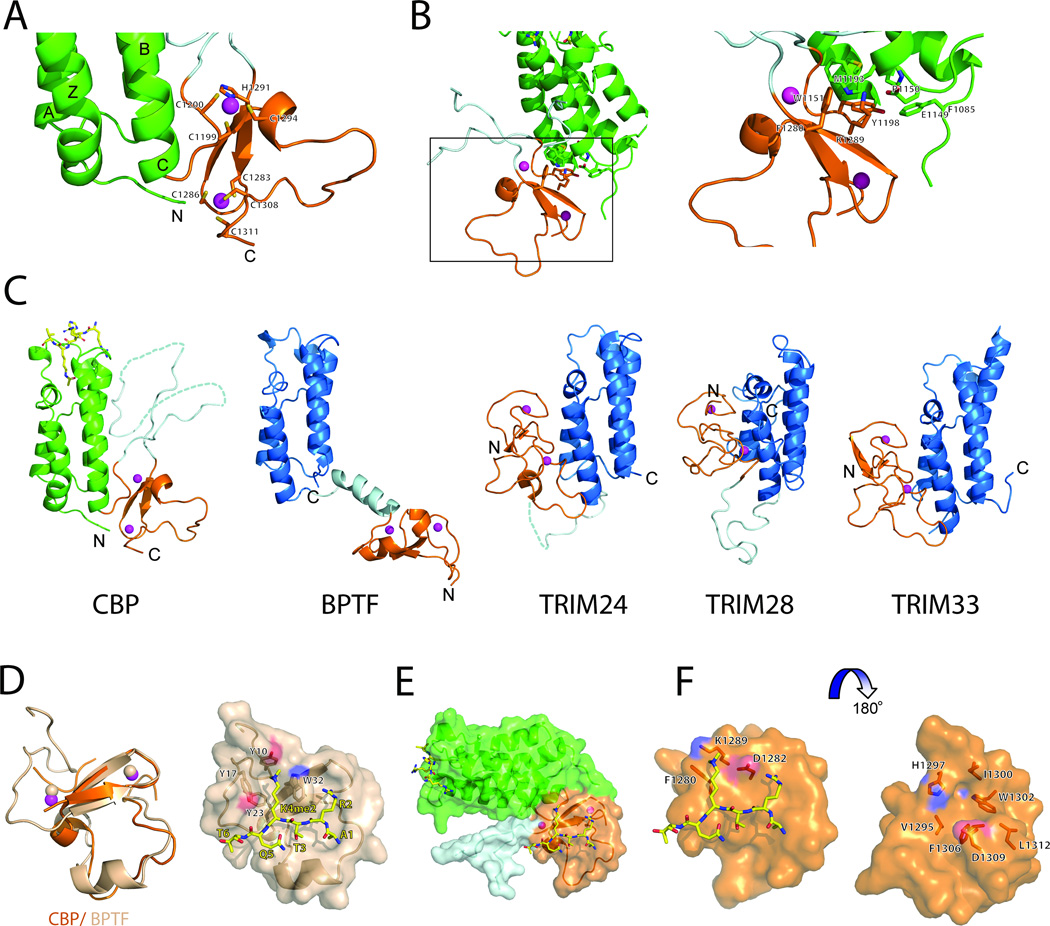
Figure 2. Molecular basis of histone recognition of the BrD-PHD module of CBP.
(A) A close-up view of the PHD finger of CBP. The residues coordinating the Zn atoms (in magenta spheres) are in sticks and labeled.
(B) Inter-domain interactions of the bromodomain and the PHD finger of CBP.
(C) Structural comparison of the tandem BrD-PHD finger modules of CBP, BPTF, TRIM24, TRIM28 and TRIM33. PHD modules are colored in orange, BrDs in blue and green for CBP, linkers between domain are colored in light cyan. Zn atoms are shown in magenta spheres.
(D) Superimposition of CBP and BPTF PHD modules. The domain represented in cartoons and colored in orange and salmon for CBP and BPTF, respectively. Zn atoms are shown in magenta and salmon spheres for CBP and BPTF, respectively. On the right, surface representation of BPTF PHD finger bound to H3K4me2 peptide (shown in sticks with carbon atom labeled in yellow). The residues comprising aromatic cage are shown in sticks.
(E) A surface representation of a model of CBP bound to H3K4me2 peptide. The orientation is as in A.
(F) Close-up view of a model of the CBP PHD finger bound to H3K4me2 peptide. The residues close to the methylated Lys and line-up the groove of the domain are shown in stick and labeled.
See also Figures S2 and S3.