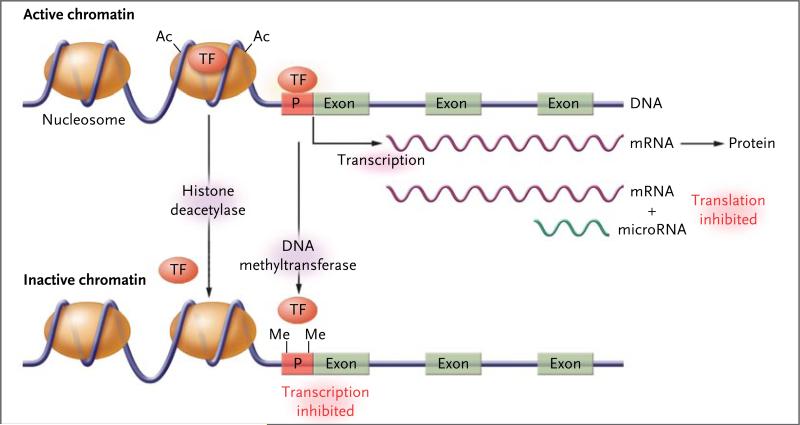
Figure 2. Regulation of Gene Expression through Epigenetic Processes.
Epigenetic modification of histones or of DNA itself controls access of transcription factors (TFs) to the DNA sequence, thereby modulating the rate of transcription to messenger RNA (mRNA). Transcriptionally active chromatin (top) characterized by the presence of acetyl groups (Ac) on specific lysine residues of core histones in the nucleosome, which decreases their binding to DNA and results in a more open chromatin structure that permits access of transcription factors. In addition, cytidine–guanosine (CpG) sequences in the promoter regions (P) of actively transcribed genes are generally unmethylated, allowing for the binding of transcription factors. Transcriptionally inactive chromatin (bottom) is characterized by histone deacetylation, promoter CpG methylation (as indicated by methyl groups [Me]), and decreased binding of transcription factors. (For simplicity, other histone modifications [such as methylation] and additional regulatory factors [such as methyl-CpG binding proteins] are not shown.) A further level of epigenetic control is provided by microRNA molecules (19 to 22 nucleotides in length), which bind to complementary sequences in the 3′ end of mRNA and reduce the rate of protein synthesis.