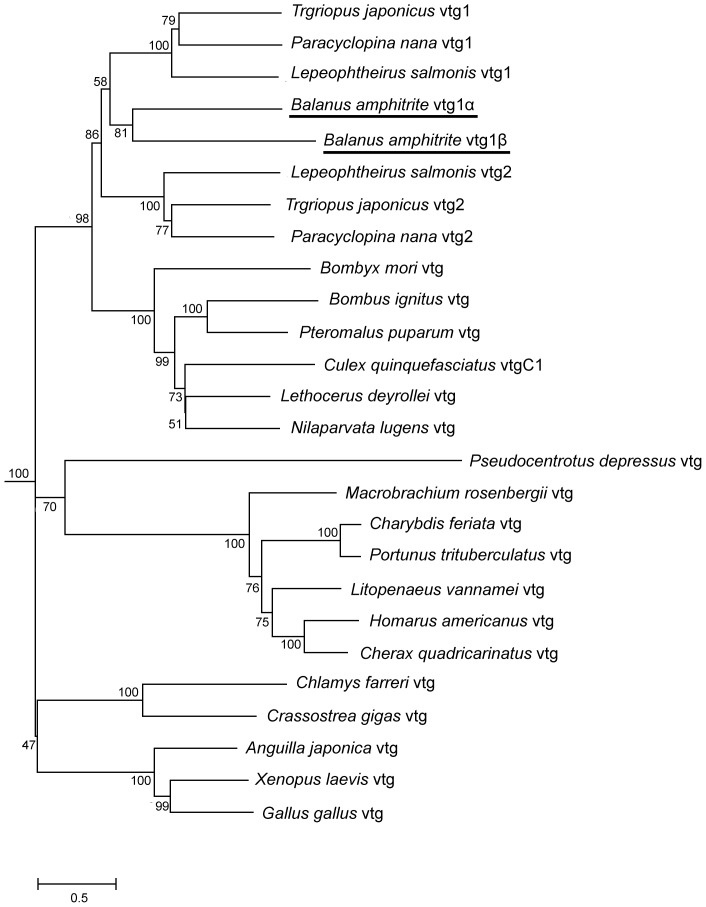
Figure 4. Neighbor-joining tree based on vitellogenin protein sequences.
Branch lengths represent substitutions per site, and numbers at each node represent bootstrap values. The sequences used were as follows: Tigriopus japonicas vtg1 (ABZ91537), Tigriopus japonicas vtg2 (ACJ12892), Paracyclopina nana vtg1 (ADD73551), Paracyclopina nana vtg2 (ADD73552), Lepeophtheirus salmonis vtg1 (ABU41134), Lepeophtheirus salmonis vtg2 (ABU41135), Bombyx mori vtg (BAA06397), Bombus ignites vtg (ACM46019), Pteromalus puparum vtg (ABO70318), Culex quinquefasciatus vtg (AAV31930), Lethocerus deyrollei vtg (BAG12118), Nilaparvata lugens vtg (AEL22916), Pseudocentrotus depressus vtg (AAK57983), Macrobrachium rosenbergii vtg (BAB69831), Charybdis feriata vtg (AAU93694), Portunus trituberculatus vtg (AAX94762), Litopenaeus vannamei vtg (AAP76571), Homarus americanus vtg (ABO09863), Cherax quadricarinatus vtg (AAG17936), Chlamys farreri vtg (ADE05540), Crassostrea gigas vtg (BAC22716), Anguilla japonica vtg (AAV48826), Gallus gallus vtg (AAA49139.1), and Xenopus laevis vtg (AAA49982).