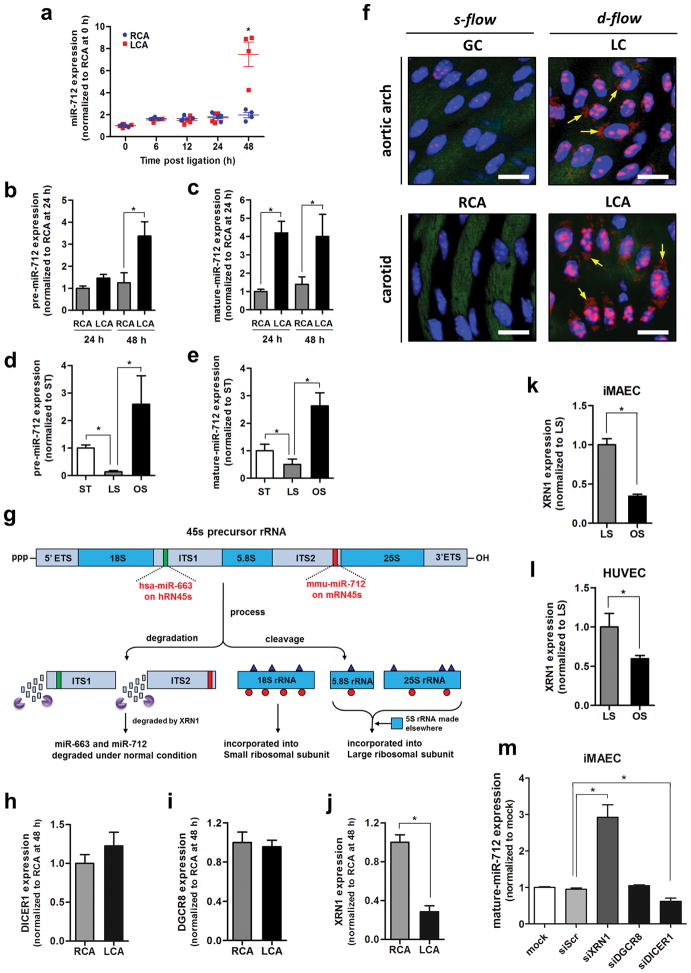
Figure 2. miR-712 is atypically derived from RN45S gene in XRN-1-dependent manner.
(a) Expression of miR-712 was determined by qPCR using endothelial-enriched RNA obtained from LCA and RCA following partial carotid ligation in C57Bl6 mouse (0–48h) (n=4, data shown as mean ± s.e.m; * p<0.05 as determined by paired t-test). (b, c) Expression of pre-miR-712 and mature miR-712 by d-flow in LCA and RCA endothelium following partial ligation at 24 and 48 hours as above in (b) was quantitated by miScript miRNA qPCR assay (n=8 each; *p<0.05 as determined by Student’s t-test). (d, e) Expression of pre-miR-712 and mature miR-712 was measured by miScript miRNA qPCR in immortalized mouse aortic endothelial cells (iMAECs) exposed to laminar (LS), oscillatory shear (OS) or static (ST) for 24 h (n=6, data shown as mean ± s.e.m; * p<0.05 as determined by Student’s t-test). (f) Aortic arch (LC and GC) and LCA and RCA obtained at 2-days post ligation obtained from control C57Bl6 mice were subjected to fluorescence in situ hybridization using digoxigenin-labeled miR-712 probe and anti-digoxigenin antibody, which was detected by tyramide signal amplification method using Cy-3 and confocal microscopy (shown in red), (n=6). Blue: DAPI nuclear stain; Green: auto-fluorescent elastic lamina; Arrows indicate cytosolic miR-712 expression. White scale bars = 20 μm. (g) shows the potential structure and processing of pre-ribosomal RNA gene, RN45s, which is composed of 18S, 5.8S and 28S rRNA sequences with 2 intervening spacers ITS1 and ITS2. The sequences matching murine miR-712 in ITS2 and its putative human counterpart miR-663 in ITS1 are indicated as well. (h–j) Expression of DICER, DGCR8 and XRN1 in mouse RCA and LCA (2-day post ligation) were determined by qPCR (n=4, data shown as mean ± s.e.m; * p<0.05 as determined by paired t-test). (k, l) XRN1 expression in iMAECs and HUVECs exposed to LS or OS for 24 h was determined by qPCR (n=3 each, data shown as mean ± s.e.m; * p<0.05 as determined by Student’s t-test). (m) miR-712 expression was induced by treating iMAECs with XRN1 siRNA but not by DGCR8 siRNA and DICER1 siRNA (n=3 each, data shown as mean ± s.e.m; * p<0.05 as determined by Student’s t-test).