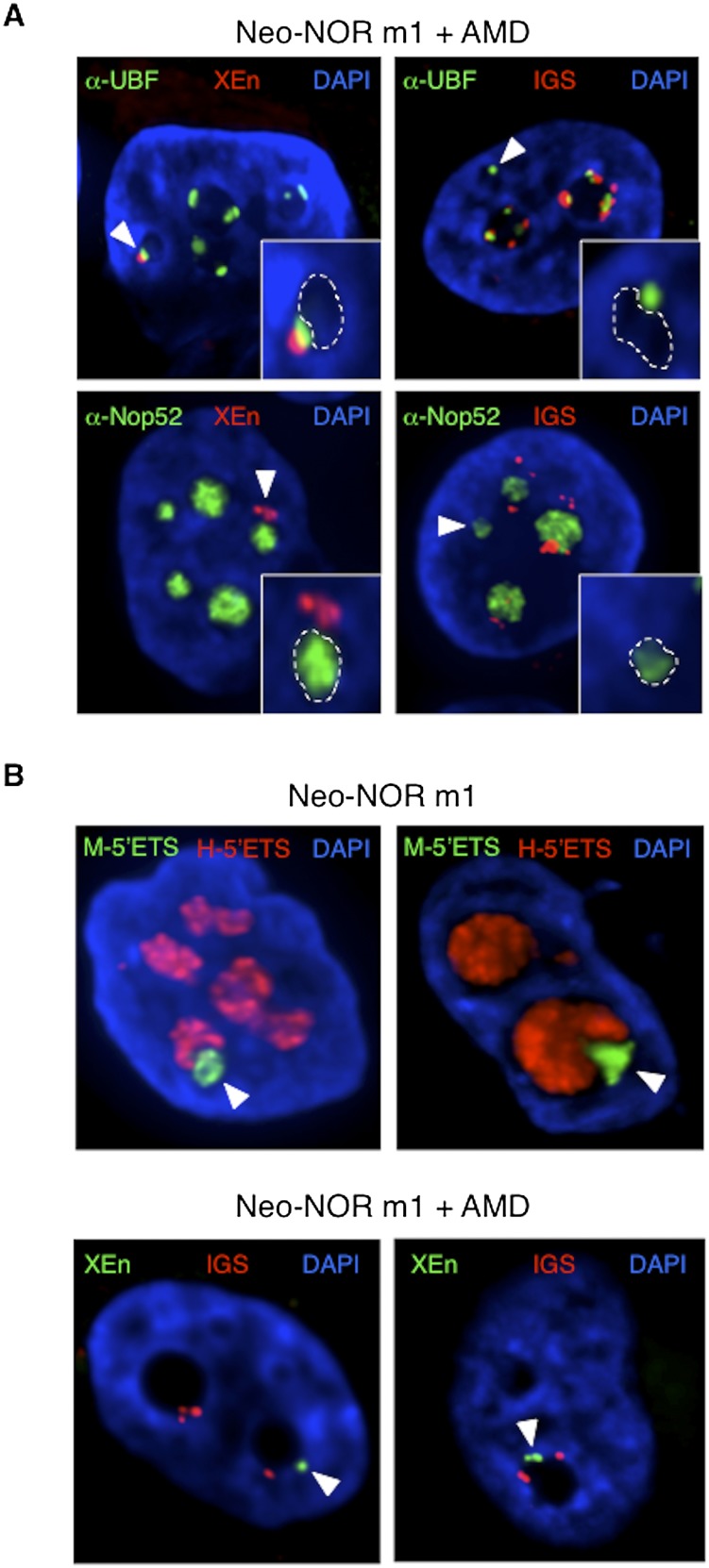
Figure 6.
Compartmentalized neonucleoli occupy distinct territories when integrated into endogenous nucleoli. (A) Combined 3D immuno-FISH performed on AMD-treated clone m1 reveals the presence of appropriately compartmentalized neonucleoli derived entirely from the neo-NOR. The top panels show UBF immunostained FC/DFC caps containing neo-NOR DNA (XEn probe; left panel) and lacking endogenous rDNA (IGS probe; right panel). The bottom panels show neonucleoli with a Nop52 immunostained GC associated with neo-NOR DNA (XEn probe; left panel) but lacking endogenous rDNA (IGS probe; right panel). Neonucleoli are indicated by arrowheads. In the enlarged insets, GCs, identified by low DAPI staining, are highlighted with a dashed line. (B, top panel) RNA-FISH performed on metacentric clone m1 identifies a distinct subnucleolar neo-NOR territory. Neo-NOR and endogenous transcripts are detected with mouse and human 5′ ETS probes, respectively. (Bottom panel) FISH performed on neo-NOR clone m1 indicates that neo-NORs and endogenous NORs are resolved into distinct nucleolar caps associated with the same GC upon AMD treatment. Neo-NORs and endogenous NORs were identified using XEn and IGS probes, respectively. GCs are identified by low DAPI staining. Images presented in the bottom panel correspond to single optical planes selected from deconvolved Z stacks.