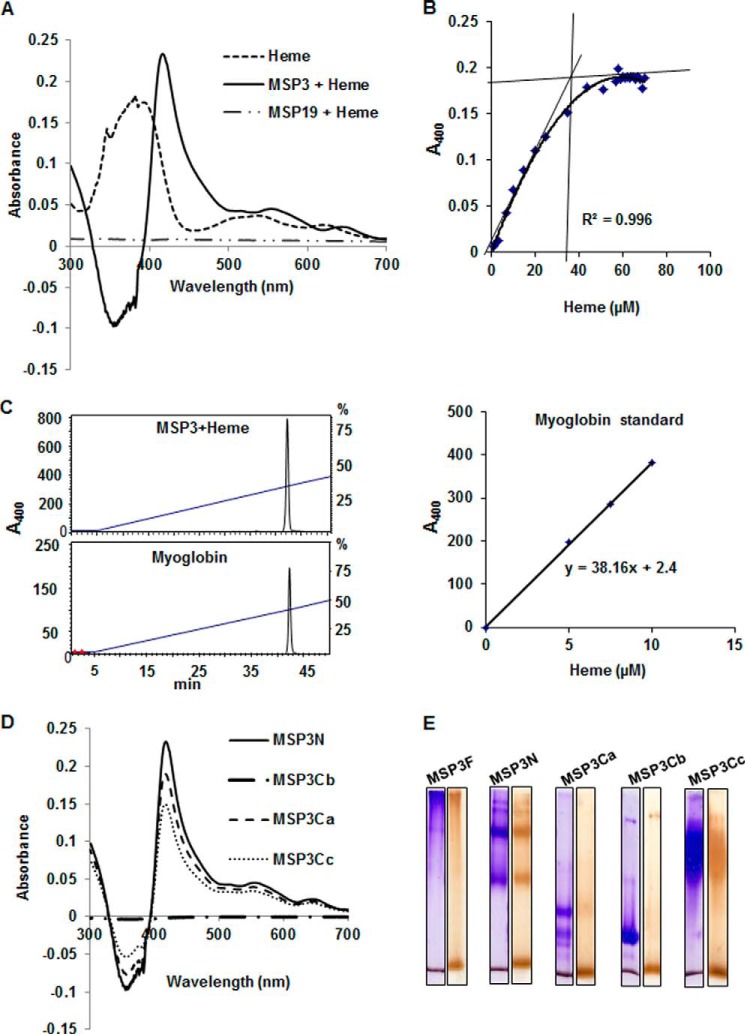
FIGURE 4.
MSP3 can bind several molecules of heme in vitro. A, UV spectrophotometry difference spectrum showed significant red shift of Soret peak of heme at 417 nm from 383 nm when MSP3F is scanned in the presence of 10 μm heme. MSP1(19) used as a control does not show absorption spectrum at 415 nm indicating absence of heme binding. B, determination of number of heme-binding sites on MSP3F by difference spectra recorded from heme titration assay and heme binding curve generated by plotting ΔA417 nm versus heme concentration showed 1:35 stoichiometry of MSP3 and heme. C, HPLC-based characterization of the MSP3F-bound heme using horse heart myoglobin and pure hemin as controls showed 1:40 stoichiometry of MSP3 and heme. D, UV spectrophotometry difference spectra showed significant red shift of Soret peak of heme at 415 nm from 383 nm when MSP3 fragments MSP3N, MSP3Cb, and MSP3Cb were recorded in the presence of 10 μm heme. MSP3Cb did not show any red shift in the presence of heme suggesting the absence of interaction. E, native PAGE analysis of heme binding by peroxidase staining. MSP3F and other polypeptides show heme interaction as evident by brown bands in peroxidase-stained native PAGE as compared with Coomassie-stained PAGE run in parallel. MSP3Cb did not show any significant binding to heme as evident by absence of brown band in peroxidase staining compared with Coomassie gel. Unbound heme is stained at the bottom of all the gel lanes.