FIGURE 1.
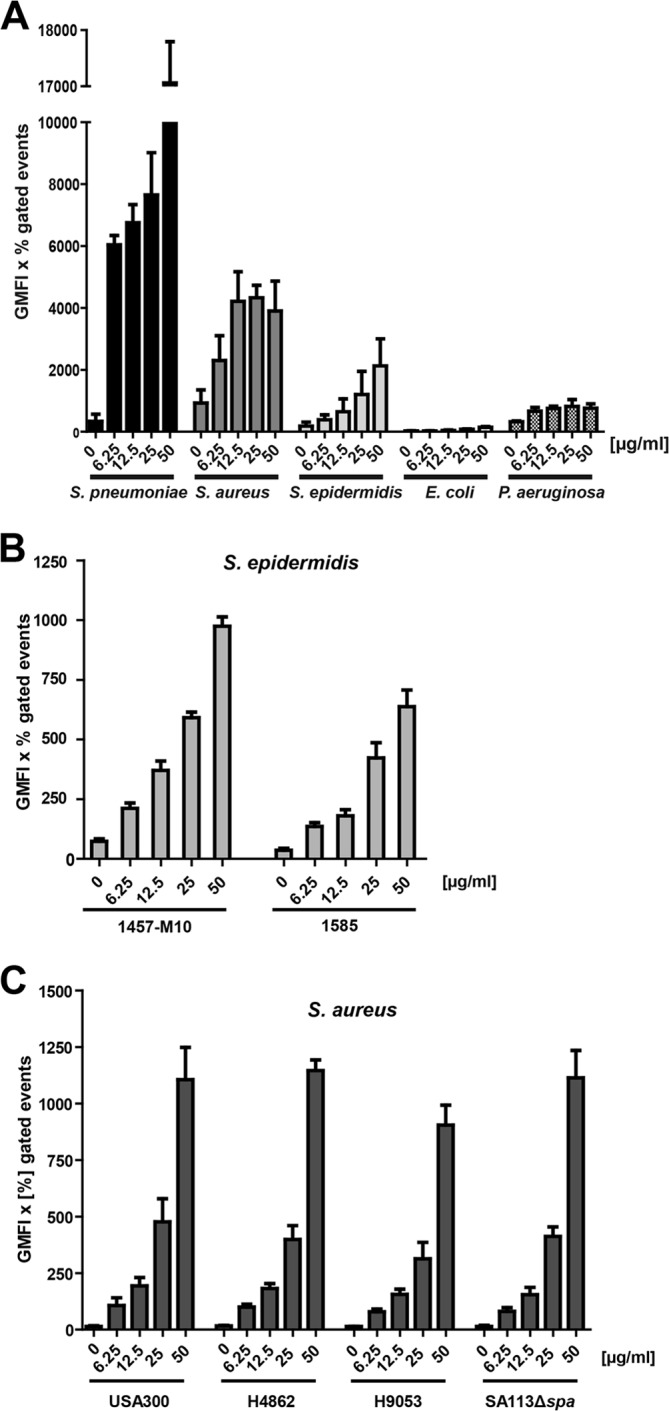
Binding of soluble hTSP-1 by bacteria. A, dose-dependent binding of hTSP-1 to Gram-positive S. pneumoniae D39Δcps, S. aureus SA113Δspa, S. epidermidis RP62A, and Gram-negative E. coli 536 and P. aeruginosa strain 6, respectively, was analyzed by flow cytometry. Bacterial suspensions of 5 × 108 bacteria in 100 μl of PBS were incubated with increasing concentrations of hTSP-1 (0–50 μg/ml) and analyzed for surface-bound hTSP-1 using a specific mouse anti-TSP-1 antibody and secondary Alexa Fluor 488 conjugated anti-mouse IgG. The values are represented as the geometrical mean fluorescence intensity multiplied with the percent of gated events (GMFI × % gated events). B, dose-dependent binding of hTSP-1 to clinical isolates S. epidermidis 1457-M10 (pia-negative mutant of a 1457 catheter infection isolate) and S. epidermidis 1585 (liquor shunt infection isolate). The values are represented as the geometrical mean fluorescence intensity multiplied with percent gated events. C, dose-dependent binding of FITC-labeled hTSP-1 to different clinical isolates of S. aureus (H4862 and H9053, furuncle isolates, and caMRSA USA300). Bacterial suspensions of 5 × 108 bacteria in 100 μl of PBS were incubated with increasing concentrations of FITC-labeled hTSP-1 (0–50 μg/ml) and analyzed for surface-bound hTSP-1 using flow cytometry. The values are represented as the geometrical mean fluorescence intensity multiplied with percent gated events.
