Background: Cappuccino is a formin actin nucleator that regulates cytoskeletal organization during Drosophila oogenesis.
Results: Cappuccino binds microtubules through two domains and cannot nucleate actin filaments when bound to microtubules.
Conclusion: Actin filament assembly and microtubule binding are mutually exclusive activities of Cappuccino.
Significance: We provide mechanistic insight into the role of formins as coordinators of the actin and microtubule cytoskeletons.
Keywords: Actin, Cell Polarity, Cytoskeleton, Formin, Microtubules, Oocyte, Capu, TIRF
Abstract
Formin family actin nucleators are potential coordinators of the actin and microtubule cytoskeletons, as they can both nucleate actin filaments and bind microtubules in vitro. To gain a more detailed mechanistic understanding of formin-microtubule interactions and formin-mediated actin-microtubule cross-talk, we studied microtubule binding by Cappuccino (Capu), a formin involved in regulating actin and microtubule organization during Drosophila oogenesis. We found that two distinct domains within Capu, FH2 and tail, work together to promote high-affinity microtubule binding. The tail domain appears to bind microtubules through nonspecific charge-based interactions. In contrast, distinct residues within the FH2 domain are important for microtubule binding. We also report the first visualization of a formin polymerizing actin filaments in the presence of microtubules. Interestingly, microtubules are potent inhibitors of the actin nucleation activity of Capu but appear to have little effect on Capu once it is bound to the barbed end of an elongating filament. Because Capu does not simultaneously bind microtubules and assemble actin filaments in vitro, its actin assembly and microtubule binding activities likely require spatial and/or temporal regulation within the Drosophila oocyte.
Introduction
Coordination of actin and microtubule cytoskeletal networks is required for a diverse set of cellular processes, from cell motility and morphogenesis to intracellular transport and nuclear migration (1, 2). Recently, formin family actin nucleators have emerged as coordinators of the actin and microtubule cytoskeletons. In addition to nucleating actin filaments, all formins tested to date directly bind microtubules in vitro (3, 4).
Although actin polymerization by formins has been well characterized, how formins coordinate actin and microtubule networks remains an open question. In fact, only three formins (mDia1, mDia2, hINF2) representing just two of the 15 metazoan formin groups have been characterized in any biochemical detail (4), and experimental data for formin-microtubule binding vary widely across the few formins studied. Several in vitro experiments suggest that actin and microtubules directly compete for formin binding: for example, microtubules potently inhibit actin polymerization by mDia2, whereas actin monomers compete with microtubules for binding to hINF2 (4). In contrast, other findings support a role for formins in the coordination of actin and microtubule networks: overexpression of mDia1 or FHOD1 aligns actin and microtubule networks in cells (5, 6), and hINF2 and Cappuccino (Capu)2 can cross-link actin and microtubules in vitro (4, 7). Although this variety likely reflects the specialized cellular roles of distinct formins, it is unclear whether conserved microtubule binding mechanisms exist or whether this functional diversity stems from fundamental differences in formin-microtubule interactions. Addressing this question will require careful biochemical analysis of microtubule binding by a variety of formins representing the many formin groups.
To further improve our mechanistic understanding of formin-microtubule interactions and formin-mediated actin-microtubule cross-talk, we studied microtubule binding by the Drosophila formin Capu. Capu belongs to the FMN group of formin nucleators and has two conserved mammalian homologs, Fmn-1 and Fmn-2 (8). All three proteins are involved in cytoskeletal regulation during key stages of development. Capu is required for establishing the major body axes of the developing Drosophila oocyte, and loss of Capu leads to severe polarity defects and female sterility (9). Similarly, Fmn-1 and Fmn-2 have been implicated in a variety of developmental processes, including limb patterning and oocyte spindle positioning (10–12). Although these proteins all bind microtubules in vitro (13, 14), little else is known about how FMN group formins interact with microtubules and coordinate the actin and microtubule cytoskeletons. The absence of comprehensive mechanistic data for these and other formins has made it difficult to study the physiological relevance of such formin-microtubule interactions.
In this study, we have characterized Capu-microtubule binding in detail and examined the relationship between the microtubule binding and actin assembly activities of Capu. We report that Capu binds microtubules with high affinity, suggesting that this is a physiologically relevant interaction. Binding requires distinct residues within the formin homology 2 (FH2) domain as well as nonspecific charge-based interactions with the C-terminal tail domain of Capu. Additionally, we found that Capu does not bind microtubules and assemble new actin filaments simultaneously. Specifically, microtubules are potent inhibitors of Capu's actin nucleation activity both in the absence and presence of profilin, but cannot effectively compete for binding to Capu that is already bound to the barbed end of an elongating actin filament. This study provides new mechanistic information about formin-microtubule binding and represents the first detailed biochemical study of microtubule binding by a FMN group formin. Together with biochemical studies of other formins, our findings offer insight into the physiological relevance of the formin-microtubule interaction and will help create necessary tools for future in vivo work.
EXPERIMENTAL PROCEDURES
DNA Constructs and Capu Mutant Cloning
Wild-type (amino acids 467–1059), mutant, and truncated CapuCT constructs were subcloned into a modified version of pET15b with an N-terminal His6 tag as described previously (15). The Capu tail (amino acids 1029–1059) was subcloned into the vector pGEX-6P-2 (GE Healthcare) as described (15). Point mutations were introduced using QuikChange site-directed mutagenesis (Stratagene).
To identify the D854N point mutation, ovaries were dissected from capu2L-7-13 females (16) in cold ionically matched adult Drosophila saline buffer (17). RNA was isolated from the ovaries using the RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions for animal tissue samples. cDNA was generated using the Maxima Universal First Strand cDNA synthesis kit (Fermentas), subcloned into the vector pJET1.2 (Fermentas), and sequenced (GENEWIZ).
Protein Expression and Purification
Acanthamoeba castellani actin, chickadee (Chic; Drosophila profilin), and Schizosaccharomyces pombe profilin were purified according to published protocols (18, 19). All His6-tagged CapuCT and CapuFH2 proteins were expressed and purified as described (15) with the following modifications for CapuFH2: dialysis buffers were supplemented with 50 mm KCl and storage buffer contained 100 mm NaCl to improve protein solubility. The total concentration of protein was calculated by quantitative Sypro-Red (Invitrogen) staining using wild-type CapuCT as a standard.
GST and GST-tail were expressed as described (15). Briefly, extracts were passed over glutathione-Sepharose (GE Healthcare), eluted using glutathione, dialyzed overnight against 10 mm Tris-HCl, pH 8.0, 1 mm DTT, and further purified over a MonoQ anion exchange column (GE Healthcare).
Microtubule Preparation
Frozen aliquots of tubulin in BRB buffer (80 mm K-PIPES, pH 6.8, 1 mm EGTA, 1 mm MgCl2) supplemented with 1 mm GTP were prepared from lyophilized tubulin (catalog no. T240, Cytoskeleton, Inc.). Tubulin was thawed quickly at 37 °C, incubated for 5 min on ice, and centrifuged at 100,000 × g for 10–20 min at 4 °C. Tubulin concentration in the supernatant was determined by its absorbance at 280 nm (ϵ280 = 115,000 cm−1 m−1) and subsequently polymerized in BRB buffer plus 2 mm GTP at 37 °C by addition of 0.1, 1, and 20 μm taxol (paclitaxel, Sigma-Aldrich) at 10-min intervals from 100-fold concentrated dimethyl sulfoxide stocks.
For experiments with subtilisin-treated microtubules, polymerized tubulin was incubated with 100 μg/ml subtilisin (Sigma-Aldrich) at 37 °C for 1 h. The reaction was quenched with 2 mm PMSF, and microtubules were spun over a 10% sucrose cushion at 70,000 × g for 10 min at 25 °C to remove cleaved peptides. The pellet was washed and resuspended in microtubule-binding buffer (MTB; 10 mm Na-HEPES, pH 7.0, 1 mm EGTA, 1 mm MgCl2, 1 mm DTT, 20 μm taxol, 0.5 mm thesit (nonaethylene glycol monodecyl ether, P-9641; Sigma-Aldrich) and 50 or 100 mm KCl as indicated).
Microtubule-binding Assays
Microtubule-binding assays were performed essentially as described (13) with modifications. Briefly, CapuCT proteins, GST, or GST-tail were cleared at 100,000 × g for 20 min at 4 °C. For actin monomer competition experiments, actin was preincubated on ice with a 2-fold molar excess of latrunculin B (LatB; EMD Millipore Chemicals). LatB-actin and Chic samples were each diluted in MTB and cleared at 100,000 × g for 20 min at 4 °C. Protein concentration in the supernatant was determined by absorbance at 280 nm or 290 nm for LatB-actin. Proteins were mixed with microtubules (0.5 μm) or buffer in MTB. Samples were incubated for 15 min and centrifuged at 100,000 × g for 10 min at 25 °C. Supernatants were removed, and pellets were washed before resuspending in SDS-PAGE sample buffer.
Proteins were resolved by SDS-PAGE, stained with Sypro-Red (Invitrogen), and quantified using a Pharos FX Plus Molecular Imager with Quantity One software (Bio-Rad). Signals from samples without microtubules were subtracted from the corresponding signals from samples containing microtubules to account for nonspecific protein pelleting. Protein and tubulin concentrations in each pellet sample were quantified using a standard curve for tubulin and the protein in question generated on the same gel. All concentrations reported are dimer concentrations of CapuCT, GST, or tubulin.
Binding data were fit to the McGhee von Hippel model for binding to a polar one-dimensional lattice (20),
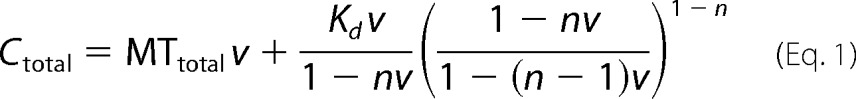 |
where Kd = e−ΔrG/RT.
The binding density v was measured by dividing the concentration of bound protein by the total concentration of polymerized tubulin dimers in the pellet (MTtotal). For large Ctotal, the value of v approaches a maximum value of 1/n where n represents the number of tubulin dimers per binding site. Because we can arbitrarily use the dissociation constant Kd versus the association constant Ka of the binding reaction, we instead fit the free energy (ΔrG) of the binding reaction, as this is unbiased by the direction of the reaction. We then converted ΔrG to the Kd values reported in Table 1. The fit parameters ΔrG and n were determined using the Levenberg-Marquardt algorithm implemented in Python using SciPy software (43).
TABLE 1.
Fit parameters for all microtubule binding curves
The free energy of binding (ΔrG) is expressed with units kJ/mol and the binding site size (n) is expressed as the number of α/β-tubulin dimers per binding site. Error bars for ΔrG and n represent one S.D. The most probable dissociation constant (Kd) values are reported in units of μm and calculated from ΔrG at 25 °C (see “Experimental Procedures” for additional information). Experiments performed with high salt conditions (100 mm KCl) and/or subtilisin-treated microtubules (sMT) are indicated; all other binding curves were measured at 50 mm KCl with untreated microtubules.
| Binding Experiment | ΔrG | Kd | n (α/β dimers) |
|---|---|---|---|
| kJ/mol | μm | ||
| CapuCT | 5.21 ± 0.16 | 0.12 | 1.44 ± 0.01 |
| CapuCT + 100 mm KCl | 0.55 ± 0.20 | 0.80 | 1.67 ± 0.04 |
| CapuCT + sMT | 1.47 ± 0.46 | 0.55 | 1.89 ± 0.09 |
| CapuCT + sMT + 100 mm KCl | −5.67 ± 0.33 | 9.8 | 2.30 ± 0.31 |
| GST-tail | −0.26 ± 0.14 | 1.1 | 0.83 ± 0.01 |
| GST-tail + 100 mm KCl | −5.37 ± 0.15 | 8.7 | 1.40 ± 0.07 |
| GST-tail + sMT | −3.55 ± 0.40 | 4.2 | 1.43 ± 0.11 |
| GST-tail + sMT + 100 mm KCl | −4.58 ± 0.29 | 6.4 | 2.94 ± 0.16 |
| CapuCT-Scr | 6.31 ± 0.58 | 0.08 | 1.40 ± 0.04 |
| CapuCT-L1048A | 5.56 ± 0.10 | 0.11 | 1.35 ± 0.01 |
| CapuCT-I706A | 4.71 ± 0.25 | 0.15 | 1.41 ± 0.02 |
| CapuCT-D854N | 5.66 ± 0.29 | 0.10 | 1.44 ± 0.02 |
| CapuCT-K858A | 4.01 ± 0.17 | 0.20 | 1.20 ± 0.01 |
| CapuCT-K856A | 3.33 ± 0.11 | 0.26 | 0.83 ± 0.01 |
Pyrene-actin Polymerization Assays
Pyrene-actin assembly assays were carried out as described (19). For the experiments in Fig. 2, buffers were supplemented with 20 μm taxol from a 10 mm stock in dimethyl sulfoxide. Unless stated otherwise, microtubules were added to the polymerization buffer before addition to magnesium-charged monomeric actin. For experiments with profilin, S. pombe profilin was briefly pre-incubated with monomeric actin and used at a final concentration of 8 μm. Fluorescence was monitored in a spectrofluorometer (Photon Technology) or a TECAN F200 with λexcitation = 365 nm and λemission = 407 nm.
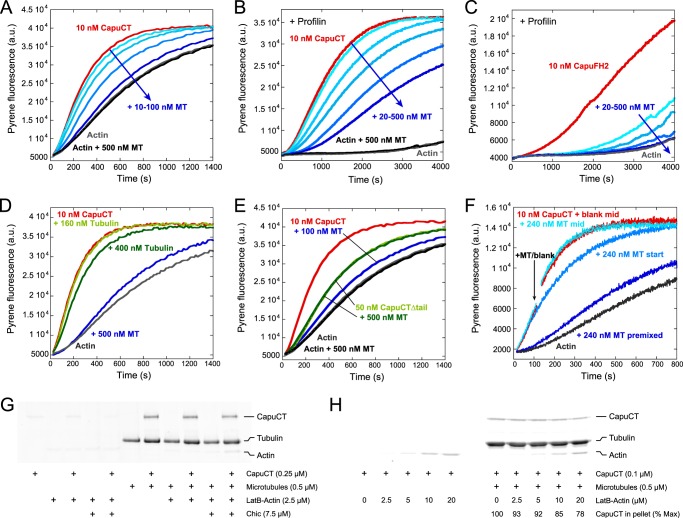
FIGURE 2.
Microtubules potently inhibit nucleation activity of Capu. A, dose-dependent inhibition of 10 nm CapuCT by 10, 20, 50, and 100 nm microtubules (MT) in bulk pyrene-actin polymerization assays. Microtubules do not affect baseline actin polymerization rates. B and C, microtubules inhibit nucleation from profilin-actin by 10 nm CapuCT (B; 20, 50, 100, 200, and 500 nm microtubules) and 10 nm CapuFH2 lacking the FH1 domain (C; 20, 50, 100, 500 nm microtubules). D, unassembled tubulin dimers have a negligible effect on CapuCT actin polymerization activity compared with taxol-stabilized microtubules. E, microtubules do not inhibit actin polymerization by CapuCTΔtail. At a 10:1 molar ratio, microtubules potently inhibit actin polymerization by CapuCT but not CapuCTΔtail. F, inhibition is sensitive to the time at which microtubules are added to the assay. Microtubules were added to CapuCT before adding actin (premixed), immediately after adding actin (start), or ∼120 s into the assay (mid, arrow indicates beginning of mixing; a.u., arbitrary units). G and H, LatB-stabilized actin monomers (LatB-Actin; 2-fold molar excess of LatB) weakly compete with microtubules to bind CapuCT in high-speed pelleting assays. Pellet fractions are shown, and concentrations are given. G, no noticeable competition was observed with 2.5 μm LatB-actin in the absence or presence of Chic (Drosophila profilin). H, some competition between microtubules and LatB-actin can be observed at very low CapuCT and very high LatB-actin concentrations, similar to those used in bulk pyrene-actin nucleation assays. CapuCT bound to microtubules is reported as a percentage of the maximum CapuCT bound at 0 μm LatB-actin.
Total Internal Reflection Fluorescence Microscopy
Cover glass was silanized and functionalized with biotin-PEG as described (19, 22). Actin was labeled at Cys-374 with OregonGreen 488 iodoacetamide (Invitrogen) or EZ-link maleimide-PEG2-biotin (Thermo Scientific) essentially as described (19). Because labeling with biotin at Cys-374 caused F-actin to depolymerize, the actin was subsequently centrifuged at 195,000 × g, and the supernatant was further purified by size exclusion chromatography on a Superdex 200 10/300 GL (GE Healthcare) in G-buffer (2 mm Tris-Cl, pH 8.0, 0.1 mm CaCl2, 0.2 mm ATP, 0.5 mm TCEP, 0.04% sodium azide). Biotin-actin could incorporate into actin filaments at low labeling fractions (<2%).
Microtubules were prepared as described above, with the exception that frozen tubulin aliquots contained 20% HiLyte647 tubulin (catalog no. TL670M Lot: 013, ∼0.2 dyes per tubulin dimer, Cytoskeleton, Inc.) and 10% biotin tubulin (catalog no. T333P Lot: 013, ∼1–2 biotins per tubulin dimer, Cytoskeleton, Inc.). F-actin seeds were polymerized from 99.4% unlabeled actin, 0.6% biotinylated actin, and equimolar Alexa Fluor 488 phalloidin (Invitrogen). Prior to imaging, ∼15-μl flow cells were built using double-stick tape. Each flow cell was prepared with the following series of washes: 25 μl 1% Pluronic-F127 (Sigma); 100 μg/ml κ-casein (Sigma) in phosphate-buffered saline for 2 min; 25 μl of TIRF buffer (KMEH (50 mm KCl, 10 mm Na-HEPES, pH 7.0, 1 mm EGTA, 1 mm MgCl2), 50 mm DTT, 20 mm glucose, 0.2 mm ATP, 0.2% methylcellulose (400 cP, Sigma)); 25 μl of 40 nm streptavidin (VWR) in KMEH for 1 min; 25 μl of TIRF buffer supplemented with 20 μm taxol, 25 μl of 5–50 nm F-actin seeds, and ∼0.5 μm taxol-stabilized microtubules in KMEH with 40 μm taxol for 2 min; and 25 μl of TIRF buffer supplemented with 20 μm taxol. The actin mix (0.6 μm actin, 30% Oregon Green 488-labeled, 3 μm Chic, ± 4 nm CapuCT, TIRF buffer, 20 μm taxol, 500 μg/ml glucose oxidase, 100 μg/ml catalase, 500 μg/ml κ -casein) was flowed into the cell and imaged after 3–4 min of lag time. Images were collected every 10 s on a DMI6000 TIRF microscope (Leica), using a z penetration depth of 110 nm.
RESULTS
Microtubule Binding by Capu
We used high-speed taxol-microtubule pelleting assays to measure microtubule binding by the C-terminal half of Capu, CapuCT, comprising the two conserved formin homology (FH) domains and short C-terminal tail domain of Capu (see Fig. 1A for diagrams of core constructs used in this study). We fit our data to a non-cooperative, polar, one-dimensional lattice binding model (20) to determine the dissociation constant (Kd) and the number of α/β-tubulin dimers (n) in each binding site. At 50 mm KCl, CapuCT binds microtubules with affinity on the order of 100 nm and occupies a binding site of ∼1.4 tubulin dimers (Fig. 1B and Table 1).
FIGURE 1.
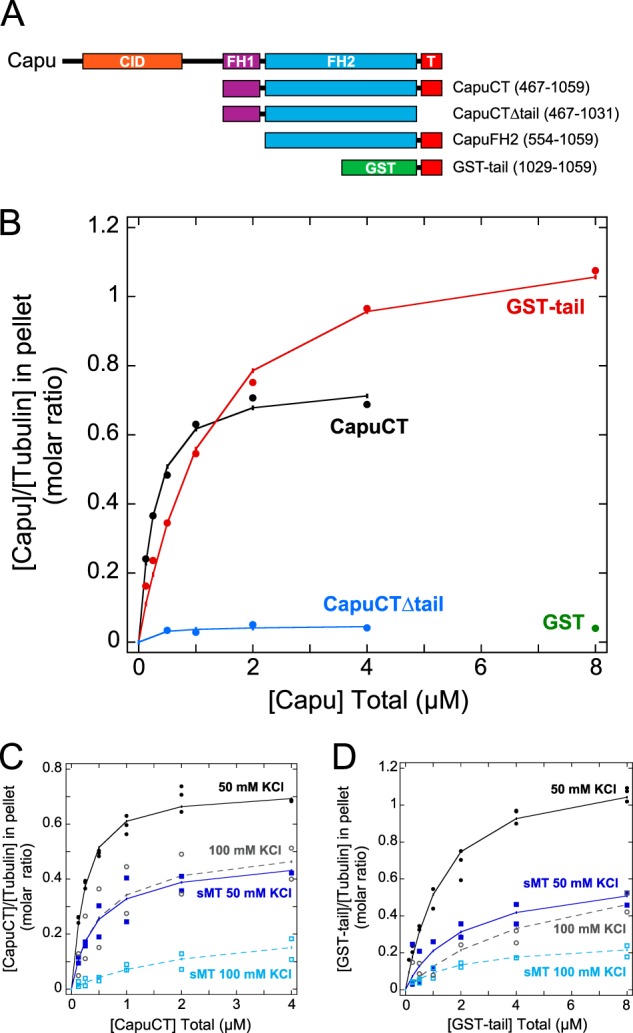
High-affinity microtubule binding requires FH2 and tail. A, schematics of Capu (1059 amino acids) and the constructs used in this paper. CID, Capu inhibitory domain (orange); FH1, formin homology-1 (purple); FH2, formin-homology-2 (blue); T, tail (red); GST (green). B, binding of CapuCT, CapuCTΔtail, GST-tail, and GST control to taxol-stabilized microtubules at 50 mm KCl. Total tubulin concentration is 0.5 μm in all binding assays. CapuCT (C) and GST-tail (D) binding to intact microtubules at 50 mm KCl (solid circles, solid line) or 100 mm KCl (open circles, dashed line) and to subtilisin-treated microtubules (sMT) at 50 mm KCl (solid squares, solid line) or 100 mm KCl (open squares, dashed line).
A previous study showed that the C-terminal tails of different formins have varying effects on microtubule binding (4). We therefore measured microtubule binding of CapuCTΔtail, a construct lacking the 30-amino acid tail domain of Capu, and found that the tail is required for microtubule binding (Fig. 1B). To test tail-microtubule binding directly, we created a GST-fusion protein of the tail domain of Capu because the orientation of the GST-tail dimer is thought to mimic the antiparallel orientation of the formin dimer (4). Although GST-tail could indeed bind microtubules, it bound with lower affinity (on the order of 1 μm) and higher binding density than CapuCT (∼0.8 tubulin dimers per binding site; Fig. 1B and Table 1). GST binding to microtubules was negligible even at the highest protein concentration used in these assays (Fig. 1B).
To further characterize the nature of Capu-microtubule binding, we assessed the contribution of ionic contacts in the microtubule pelleting assays. We first repeated the assay at a higher ionic strength. At 100 mm KCl, CapuCT and GST-tail both exhibit reduced microtubule binding density and binding affinity (Fig. 1, C and D, and Table 1). Because the Capu-microtubule interaction is sensitive to salt, we next asked whether the glutamate-rich C-terminal tails of α- and β-tubulin are involved in Capu binding. We treated microtubules with the protease subtilisin to selectively remove the acidic tubulin tails. Similar to what we observed at higher ionic strengths, both CapuCT and GST-tail exhibited reduced binding density and binding affinity to subtilisin-treated microtubules at 50 mm KCl compared with untreated microtubules (Fig. 1, C and D, and Table 1). Binding to subtilisin-treated microtubules was further reduced at 100 mm KCl (Fig. 1, C and D, and Table 1), suggesting that the tubulin tails are not the only sources of ionic interaction.
Microtubules Inhibit Actin Nucleation Activity of Capu
Because Capu is a potent actin nucleator, we next asked whether microtubule binding affects the ability of Capu to assemble actin filaments. We first performed bulk pyrene-actin polymerization assays with A. castellani actin. In these assays, Capu was preincubated with microtubules before being added to actin. In these assays, microtubules inhibited actin assembly by CapuCT in a dose-dependent manner (Fig. 2A). This effect is specific to microtubules because CapuCT was only minimally inhibited by a 40-fold molar excess of unassembled tubulin dimers (Fig. 2D). Microtubules failed to inhibit CapuCTΔtail, showing that inhibition requires high affinity microtubule binding (Fig. 2E). We used higher concentrations of CapuCTΔtail in this assay because this construct exhibits reduced polymerization activity as reported previously (15).
We next tested whether this inhibitory effect persisted when high levels of profilin were added to the assay. We used S. pombe profilin in these assays because, unlike other profilins, it exhibits minimal bias against binding actin labeled at Cys-374 (data not shown). Even in the presence of excess profilin (2:1 molar ratio with actin), microtubules inhibited CapuCT in a dose-dependent manner (Fig. 2B). Profilin not only suppresses formin nucleation activity but also works together with the FH1 domain to accelerate elongation of formin-bound filaments (23, 24). We therefore used a Capu construct lacking the FH1 domain (CapuFH2; Fig. 1A) to specifically test nucleation inhibition by microtubules in the presence of excess profilin. Although nucleation by CapuFH2 was very slow in the presence of profilin, microtubules further inhibited nucleation in a dose-dependent manner (Fig. 2C).
To test whether microtubules were hindering the elongation of Capu-associated filaments, we measured the effect of microtubules added at different timepoints throughout the polymerization assay. When we added microtubules immediately after mixing CapuCT with actin, as opposed to before actin was added, inhibition was dramatically reduced (Fig. 2F). Microtubules added midway through the assay, when nucleation is negligible, had no noticeable inhibitory effect (Fig. 2F). These results show that microtubules specifically inhibit the nucleation activity of Capu and do not noticeably slow or inhibit the elongation of Capu-associated actin filaments.
To gain a better mechanistic understanding of how microtubules inhibit the nucleation activity of Capu, we tested whether microtubules and actin monomers directly compete for CapuCT binding. We first tried the same high-speed pelleting assay conditions used by Gaillard et al. (4) to test actin monomer competition with mDia1 and hINF2. Under these assay conditions, a 10-fold molar excess of actin monomers, we did not observe a change in CapuCT binding to microtubules in the presence or absence of Drosophila profilin (Chic; Fig. 2G). However, our bulk pyrene-actin polymerization assays contain a much higher (400-fold) molar excess of actin monomers compared with CapuCT. To better simulate the conditions of our polymerization assay, we tried high-speed pelleting assays with much lower and higher CapuCT and actin concentrations, respectively. At a 200-fold molar excess of actin monomers, ∼20% of the bound CapuCT was successfully competed away from the microtubules (Fig. 2H). Together with our bulk polymerization data, these findings suggest that microtubules inhibit actin nucleation at least in part by preventing actin monomers from associating with CapuCT.
Capu Association with the Barbed End of Actin Is Not Disrupted by Microtubules
To better understand the interplay between actin polymerization and microtubule binding, we turned to single filament analysis. Using total internal reflection fluorescence (TIRF) microscopy, we directly observed elongating actin filaments in a field of immobilized, taxol-stabilized microtubules. To help limit the total number of actin filaments in these assays, filaments were grown from preassembled F-actin seeds stabilized with Alexa Fluor 488-labeled phalloidin, and CapuCT was added at very low concentrations. We included Chic to differentiate filaments with Capu bound at the barbed end from filaments free of Capu (19): in the presence of Chic, Capu-associated filaments appear dimmer and elongate much faster than bright Capu-free filaments.
In this experiment, elongating actin filaments that encountered a microtubule were observed to either cross over the microtubule and continue growing (Fig. 3A, panel i, and supplemental Movie S1); grow along the microtubule (Fig. 3A, panel ii, and supplemental Movie S2); or turn away from the microtubule and continue growing (Fig. 3A, panel iii, and supplemental Movie S3). Consistent with our bulk polymerization assays (Fig. 2F), CapuCT-associated filaments never stopped upon encountering a microtubule. The frequencies of the three observed behaviors were similar for both CapuCT-bound (dim) and CapuCT-free (bright) filaments (Fig. 3B); we therefore believe that these behaviors, although intriguing, reflect our experimental set-up. The illumination field is deep enough (110 nm) to visualize filaments crossing without physically encountering each other. At the surface of the coverslip, microtubules could act as physical barriers and cause actin filaments to turn or elongate along the microtubule. Interactions between fluorescent labels could also promote alignment of microtubules and actin filaments.
FIGURE 3.
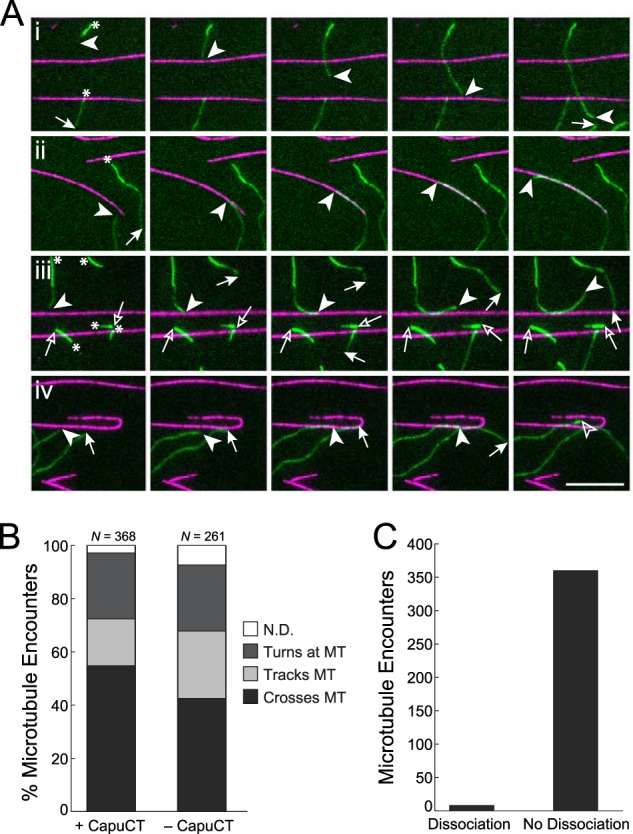
Microtubules do not disrupt Capu association with the barbed end of actin. A, upon encountering a microtubule (magenta), elongating actin filaments (green) either crossed the microtubule (i), tracked the microtubule (ii), or turned at the microtubule (iii); occasionally CapuCT dissociated from the barbed end upon encountering a microtubule (iv). Arrowheads denote the barbed ends of interest, whereas arrows denote all of the other barbed ends; free and CapuCT-associated barbed ends are represented by open and filled symbols, respectively. Asterisks in the first panel denote bright, Alexa Fluor 488-phalloidin-labeled F-actin seeds that were not nucleated by CapuCT. Images are shown at 60-s intervals. Scale bar is 10 μm. B, quantification of actin behavior upon encountering microtubules in the presence or absence of CapuCT. Events that could not be classified into one of these three categories are reported as not determined (N.D.). C, quantification of microtubule encounters that resulted in CapuCT dissociation from the barbed end of an elongating actin filament (n = 368; same data set as +CapuCT in B).
Although CapuCT occasionally dissociated from the barbed end of an actin filament upon encountering a microtubule (Fig. 3A, panel iv, and supplemental Movie S4), CapuCT remained bound to the barbed end in 98% of all of the microtubule encounters we observed (Fig. 3C). These data show directly that microtubules are unable to effectively compete CapuCT away from the barbed ends of actin filaments.
Characterization of the Tail-Microtubule Interaction
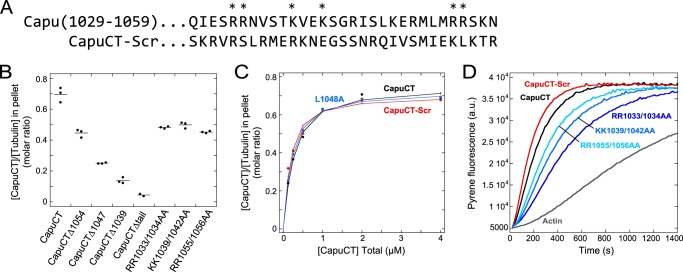
We attempted to map a more specific microtubule binding region within the tail domain. In addition to microtubule binding, the tail domain of Capu is involved in actin polymerization, protein-protein interactions, and autoinhibition (15, 19). A single point mutation, L1048A, is sufficient to disrupt Capu binding to the actin nucleator Spire (Spir) in vitro (15) but does not decrease autoinhibition (19). This mutation had no effect on microtubule binding (Fig. 4C and Table 1), suggesting that the different functions of the tail domain of Capu may be separable. We started by making a series of truncations and found that CapuCT-microtubule binding decreased monotonically as the tail was successively truncated (Fig. 4, A and B). From this, we conclude that the microtubule-binding region cannot be readily narrowed down to a small patch of neighboring residues within the tail domain.
FIGURE 4.
Tail binds microtubules through nonspecific charge-based interactions. A, sequences of wild-type and scrambled tail (residues 1029–1059). Residues mutated to alanine within wild-type are denoted by an asterisk. B, microtubule binding by CapuCT truncation and point mutation constructs. Experimental conditions are 0.5 μm tubulin and 2 μm designated CapuCT construct in 50 mm KCl buffer. Individual experiments are plotted as dots with a line showing the mean. C, CapuCT with a scrambled tail (CapuCT-Scr) or a Spir-binding point mutation (CapuCT-L1048A) bind microtubules comparably with wild-type CapuCT. All binding assays are at 50 mm KCl with 0.5 μm tubulin. D, actin polymerization activity of 10 nm each CapuCT tail point mutant and CapuCT-Scr compared with wild-type (a.u., arbitrary units).
Our earlier experiments showed that microtubule binding by Capu is sensitive to ionic strength and is therefore likely mediated by charge-based interactions. Consistent with this hypothesis, the tail domain of Capu is basic, with a theoretical pI of 11.7 (25) and a fairly even distribution of positively charged residues. To test more directly whether positively charged residues within the tail domain are important for microtubule binding, we created several arginine/lysine-to-alanine double point mutations (Fig. 4A). Despite being spread throughout the tail domain of Capu, each set of mutations had a measurable and approximately equal effect on microtubule binding (Fig. 4B). These mutations also decreased the actin nucleation activity of Capu (Fig. 4D), suggesting that Capu tail contributions to microtubule binding and actin assembly are not readily separable.
Together with our truncation results, this point mutation data suggested that the tail domain of Capu binds microtubules through nonspecific charge-based interactions. To investigate this further, we generated a CapuCT mutant in which the tail residues were randomly scrambled to give a new sequence with the same total charge and theoretical pI as the original (CapuCT-Scr; Fig. 4C, Table 1). CapuCT-Scr bound microtubules with comparable affinity and binding density to wild-type CapuCT, further supporting our model that the Capu tail binds microtubules nonspecifically. Interestingly, scrambling the tail sequence of CapuCT did not diminish the actin nucleation activity of CapuCT-Scr (Fig. 4D).
FH2 Domain Mutations Alter Microtubule Binding
Although the tail domain of Capu is required for microtubule binding, the differences in affinity and binding density between CapuCT and GST-tail indicated that the FH2 domain also contributes to microtubule binding. Because microtubules inhibit the actin polymerization activity of Capu, we first mutated two conserved actin binding residues in the FH2 domain of Capu (I706A and K856A; see Fig. 6B) that are known to abolish actin assembly activity in the yeast formin Bni1 (26). The I706A mutation dramatically reduced the actin nucleation activity of Capu but had little effect on microtubule binding (Fig. 5, A and B, and Table 1). Unexpectedly, the K856A mutation had a negligible effect on actin nucleation activity but caused a noticeable increase in microtubule binding density. The binding density of CapuCT-K856A was intriguingly similar to that of GST-tail, with an approximate binding site size of 0.8 tubulin dimers versus 1.4 for wild-type CapuCT (Fig. 5, A and B, and Table 1). Consistent with this, Capu-CT K856A is more sensitive to microtubule inhibition than wild-type CapuCT in pyrene-actin polymerization assays (Fig. 5C).
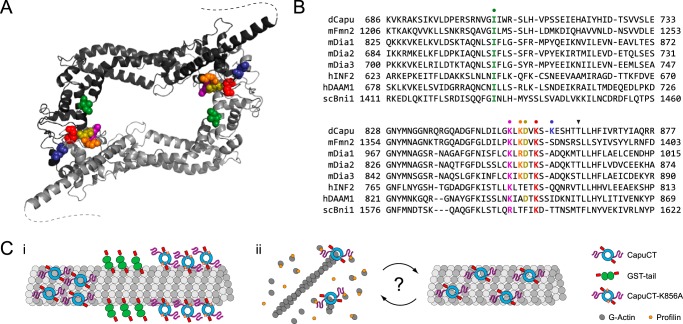
FIGURE 6.
Model of Capu-microtubule binding. A, expected locations of Capu FH2 mutations based on a Capu homology model. Residues are colored as in B, and dashed lines represent the disordered C-terminal tail domains (residues 1037–1059). Homology model was generated from the hDAAM1 crystal structure (Protein Data Bank code 2J1D (27)) using SWISS-MODEL (42). B, sequence alignments of several formins in the regions surrounding Ile-706 (green), Lys-851 (purple), Lys-853 (orange), Asp-854 (gold), Lys-856 (red), and Lys-858 (blue). Sequences were acquired from the NCBI Protein Database (Drosophila Capu, NP_476966; mouse Fmn2, NP_062318; mouse Dia1, NP_031884; mouse Dia2, NP_062644; mouse Dia3, NP_766081; human INF2, NP_071934; human DAAM1, NP_055807; Saccharomyces cerevisiae Bni1p, NP_014128) and aligned using ClustalW (21). The arrowhead denotes an mDia3 threonine residue shown previously to be phosphorylated by Aurora B kinase (31). C, schematic model for Capu-microtubule binding and actin-microtubule coordination. i, CapuCT binds microtubules through both its FH2 and tail domains; changes in binding density reflect the level of contact between the FH2 domain and the microtubule lattice. ii, actin barbed ends and microtubules compete for Capu binding; regulatory factors such as binding partners or post-translational modifications could control the degree of actin versus microtubule binding in vivo.
FIGURE 5.
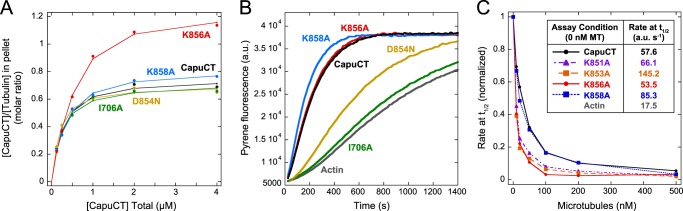
FH2 domain residues contribute to microtubule binding. A, microtubule binding by CapuCT-I706A, -D854N, -K858A, -K856A, and wild-type at 50 mm KCl with 0.5 μm tubulin. B, actin polymerization activity of 10 nm each CapuCT FH2 mutant. C, CapuCT-K856A, but not CapuCT-K858A, is more sensitive to microtubule inhibition than wild-type CapuCT. Two additional FH2 mutants, CapuCT-K851A and CapuCT-K853A, also exhibited increased sensitivity to microtubule inhibition. For each construct, 10 nm was used and rates at the time until half-maximal polymerization (t½) were normalized to the maximum polymerization rate (0 nm microtubules (MT)) and the baseline polymerization rate (actin) were reported in the Table inset (a.u., arbitrary units).
We next investigated whether other residues around Lys-856 are important for actin nucleation or microtubule binding. First, we examined a point mutation (D854N; Fig. 6B) that we identified by sequencing an EMS-generated capu mutant from an earlier screen (16). Similar to I706A, this D854N mutation had little effect on microtubule binding, but markedly decreased the actin nucleation activity of Capu (Fig. 5, A and B, and Table 1). In contrast to both D854N and K856A, mutating a poorly conserved lysine residue (K858A; Fig. 6B) had little effect on either actin nucleation activity or microtubule binding (Fig. 5, A and B, and Table 1). Two additional mutants we tested, K851A and K853A, were unstable in pelleting assays but had robust actin nucleation activity. We indirectly tested microtubule binding by these mutants by measuring sensitivity to microtubule inhibition in bulk pyrene-actin polymerization assays. Both mutants exhibited an increased sensitivity to microtubule inhibition similar to CapuCT-K856A (Fig. 5C). As a control, we also measured microtubule inhibition of CapuCT-K858A and saw very little difference compared with wild-type (Fig. 5C). From this, we concluded that microtubule binding is mediated by specific residues within the FH2 domain and, unlike binding by the tail domain, is not dependent on random charge-based interactions. Therefore, although the tail domain is required for high affinity binding, FH2 domain interactions are likely necessary to properly orient Capu on the microtubule lattice.
DISCUSSION
Microtubule Binding
Based on structural information and our experimental findings, we propose a simple model to describe the mechanism of Capu-microtubule binding (Fig. 6C). A homology model of Capu based on the crystal structure of hDAAM1 (27) reveals that residues within the FH2 domain that affect microtubule binding (Lys-851, Lys-853, Lys-856) are clustered into a positively charged patch near the base of the Capu tail domain, suggesting that these regions form a continuous binding surface (Fig. 6A). Negatively charged residues in the patch (Asp-854) or positively charged residues located away from the patch (Lys-858) have little or no effect on microtubule binding. Capu binds microtubules through a seemingly synergistic interaction involving both its tail and FH2 domains. The FH2 domain alone is insufficient to measurably bind microtubules in vitro, but the highly charged tail domain could act as an electrostatic tether that promotes FH2 binding by increasing the local FH2 concentration at the microtubule surface.
We further propose that the differences in microtubule binding density we observed reflect the size of the FH2 binding footprint along the microtubule lattice: loss (GST-tail) or reduction (K856A mutation) of FH2 domain binding results in a smaller binding footprint and a corresponding increase in binding density (Fig. 6C, panel i). This model could also explain the different microtubule binding densities seen among different formins. An emerging trend shows that formins whose tails can bind microtubules (Capu and mDia2) have higher binding densities than formins whose tails do not bind microtubules (mDia1 and hINF2) (4). In the absence of tail binding, formins may rely on more extensive FH2 contacts with the microtubule lattice and thus have a larger binding footprint.
Effect of Microtubules on Actin Assembly
Our data also support a model in which Capu does not simultaneously assemble actin filaments and bind microtubules (Fig. 6C, panel ii). We found that microtubules could potently inhibit the actin nucleation activity of Capu in bulk assays but had little effect on Capu once it was bound to the end of an elongating actin filament. The location of residues Lys-851, Lys-853, and Lys-856 on our homology model suggests that the FH2 domain binds both microtubules and the actin barbed end through similar or overlapping surfaces, further supporting our model that microtubules and actin barbed ends directly compete for Capu binding. When Capu binds microtubules, its tail domain becomes unavailable for actin monomer binding (15, 28) and the inner FH2 domain becomes sterically occluded and inaccessible to newly formed actin barbed ends. Conversely, the microtubule binding surface in the FH2 domain is inaccessible when Capu is associated with an actin barbed end, allowing Capu to elongate actin filaments in the presence of microtubules.
Our TIRF microscopy assays provide additional insight into important questions surrounding formin-mediated actin-microtubule cross-talk. Microtubules did not anchor elongating actin filaments nor act as scaffolds for actin nucleation. Although we did observe actin filaments tracking along microtubules, this behavior was essentially the same in the presence or absence of CapuCT, suggesting that low concentrations of Capu do not actively align or bundle microtubules and actin filaments. It was previously shown that Capu can cross-link actin filaments and microtubules (7), most likely through side binding of both microtubules and actin filaments. However, at the very low CapuCT concentrations used in our TIRF assays, we expect much of the CapuCT to be bound to the barbed end of elongating filaments and unavailable for binding the sides of actin filaments. Because of this, we were specifically testing the relationship between microtubule and barbed end binding rather than filament side binding.
Implications for the Role of Capu in Drosophila Oogenesis
There is strong evidence that Capu helps build a cytoplasmic actin mesh in the mid-stage Drosophila oocyte (29, 30). Here we consider the role of Capu as both a microtubule binding protein and actin assembly factor in the ooctye and how these functions might be regulated. Our observation that actin barbed ends and microtubules compete for Capu binding suggests that Capu does not simultaneously bind microtubules and assemble actin filaments within the oocyte. Acting as a microtubule binding protein, Capu could directly regulate the microtubule cytoskeleton without invoking its actin assembly activity. It could cross-link microtubules to each other and/or to pre-existing actin filaments. Conversely, when assembling actin filaments and not binding microtubules, Capu could still indirectly influence microtubule organization through the actin cytoskeleton. Spatial and/or temporal regulation of Capu could control when Capu is associated with microtubules versus actin filaments and could be achieved through additional binding partners, post-translational modification, or some combination thereof. For instance, Capu binds the actin nucleator Spir through its tail domain (15). Spir competes directly with microtubules for Capu binding (13) and forms a functional nucleation unit when bound to Capu (13, 15). This Spir-Capu nucleation unit may be much more efficient at nucleating actin in the microtubule-rich oocyte than Capu would be alone. Additional unidentified binding partners may also play a role in regulating Capu association with microtubules. Several mammalian Diaphanous family formins bind and/or colocalize with microtubule-associated proteins such as EB1, APC, and CLIP-170 (31–33). Future work will determine whether Capu has similar binding partners within the Drosophila oocyte.
Post-translational modifications, especially phosphorylation, are commonly used to regulate microtubule binding by a variety of microtubule-associated proteins (34, 35). With respect to formins, mDia3 association with microtubules has been shown to be mediated by phosphorylation by the Aurora B kinase (31). Notably, one mDia3 phosphorylation site is conserved across many formins and is within 10 amino acids of the conserved Lys-856 residue of Capu (Fig. 6B). Moreover, lysine-to-alanine mutations in this region of mDia1 have been shown to disrupt actin-microtubule coordination in Hela cells (5), suggesting that this region within the FH2 domain could be a hotspot for microtubule binding among formins. Additionally, we anticipate that post-translational modification within the tail of Capu will be important for regulating Capu function in the Drosophila oocyte. Although only ∼30 amino acids long, the tail domain is involved in Spir binding, microtubule binding, actin nucleation, and Capu autoinhibition (15, 19). Such a promiscuous domain will likely require careful regulation in vivo.
Finally, it is possible that microtubules themselves are a means of regulating Capu activity in the oocyte. Throughout mid and late oogenesis, microtubules are nucleated from all regions of the oocyte cortex except the posterior pole (36), and it was recently shown that Capu-dependent actin projections emanate specifically from the posterior cortex of the oocyte (37). Could microtubule organization restrict the location of these Capu-dependent actin projections to the oocyte posterior? Similarly, microtubules could tune the processivity of Capu as it elongates actin filaments within the oocyte. Our TIRF experiments show that microtubules do not effectively compete Capu away from the barbed end of actin filaments, causing barbed end dissociation in only 2% of all microtubule encounters. However, this relatively low probability of dissociation could have a much more substantial effect over the span of the entire oocyte where a single barbed end may encounter hundreds of microtubules. Together with the autoinhibitory activity of Capu (19), microtubules could also prevent Capu from nucleating new actin filaments after falling off the end of an elongating filament.
Concluding Remarks
Our results provide mechanistic details of Capu-microtubule binding and the interplay of microtubule binding and actin assembly in vitro. Beyond providing valuable insight into the role of Capu as a cytoskeletal regulator in the Drosophila oocyte, these findings may help advance our understanding of the Capu mammalian homologs, Fmn-1 and Fmn-2. Capu FH2 residues 851–856 are perfectly conserved in Fmn-1 and Fmn-2, and both formins contain well conserved, short, basic C-terminal tail domains, suggesting a conserved mechanism for microtubule binding (13). These formins have been implicated in a number of processes in a wide variety of cell types, including intercellular adhesion and cell spreading, as well as spindle positioning and cytokinesis during mammalian oogenesis (10, 11, 38–41). How our findings relate to the broader class of formin proteins remains to be seen. Although the microtubule binding FH2 residues we identified are well conserved across several formin groups, the C-terminal tail domains are more variable. Gaillard et al. (4) recently showed that INF2, mDia1, and mDia2 have distinct microtubule interaction properties. Notably, mDia2 has the most basic tail of the three formins and behaves the most like Capu in vitro. Future work will reveal whether our model for Capu-microtubule binding can be generalized to other formins such as mDia2.
Acknowledgments
We thank members of the Quinlan laboratory for useful discussions as well as Antonina Roll-Mecak for protocols and helpful advice. We especially thank Andrey Shur for invaluable technical help preparing proteins and reagents.
This work was supported, in whole or in part, by National Institutes of Health Ruth L. Kirschstein National Research Service Award GM007185 (to E. A. R.-J.), National Institutes of Health-National Research Service Award Fellowship F32GM087857 (to C. L. V.), National Institutes of Health, NIGMS Grant R01 GM096133, a Burroughs Welcome Fund Career Award in the Biomedical Sciences, and March of Dimes Foundation Grant 5-FY10-81 (to M. E. Q.).

This article contains supplemental Movies S1–S4.
- Capu
- Cappuccino
- FH2
- formin homology 2
- TIRF
- total internal reflection fluorescence
- Spir
- Spire
- LatB
- latrunculin B
- Chic
- Chickadee.
REFERENCES
- 1. Goode B. L., Drubin D. G., Barnes G. (2000) Functional cooperation between the microtubule and actin cytoskeletons. Curr. Opin. Cell Biol. 12, 63–71 [DOI] [PubMed] [Google Scholar]
- 2. Rodriguez O. C., Schaefer A. W., Mandato C. A., Forscher P., Bement W. M., Waterman-Storer C. M. (2003) Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat. Cell Biol. 5, 599–609 [DOI] [PubMed] [Google Scholar]
- 3. Chesarone M. A., DuPage A. G., Goode B. L. (2010) Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat. Rev. Mol. Cell Biol. 11, 62–74 [DOI] [PubMed] [Google Scholar]
- 4. Gaillard J., Ramabhadran V., Neumanne E., Gurel P., Blanchoin L., Vantard M., Higgs H. N. (2011) Differential interactions of the formins INF2, mDia1, and mDia2 with microtubules. Mol. Biol. Cell 22, 4575–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ishizaki T., Morishima Y., Okamoto M., Furuyashiki T., Kato T., Narumiya S. (2001) Coordination of microtubules and the actin cytoskeleton by the Rho effector mDia1. Nat. Cell Biol. 3, 8–14 [DOI] [PubMed] [Google Scholar]
- 6. Gasteier J. E., Schroeder S., Muranyi W., Madrid R., Benichou S., Fackler O. T. (2005) FHOD1 coordinates actin filament and microtubule alignment to mediate cell elongation. Exp. Cell Res. 306, 192–202 [DOI] [PubMed] [Google Scholar]
- 7. Rosales-Nieves A. E., Johndrow J. E., Keller L. C., Magie C. R., Pinto-Santini D. M., Parkhurst S. M. (2006) Coordination of microtubule and microfilament dynamics by Drosophila Rho1, Spire and Cappuccino. Nat. Cell Biol. 8, 367–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Higgs H. N., Peterson K. J. (2005) Phylogenetic analysis of the formin homology 2 domain. Mol. Biol. Cell 16, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Manseau L. J., Schüpbach T. (1989) cappuccino and spire: two unique maternal-effect loci required for both the anteroposterior and dorsoventral patterns of the Drosophila embryo. Genes Dev. 3, 1437–1452 [DOI] [PubMed] [Google Scholar]
- 10. Azoury J., Lee K. W., Georget V., Rassinier P., Leader B., Verlhac M. H. (2008) Spindle positioning in mouse oocytes relies on a dynamic meshwork of actin filaments. Curr. Biol. 18, 1514–1519 [DOI] [PubMed] [Google Scholar]
- 11. Schuh M., Ellenberg J. (2008) A new model for asymmetric spindle positioning in mouse oocytes. Curr. Biol. 18, 1986–1992 [DOI] [PubMed] [Google Scholar]
- 12. Zhou F., Leder P., Zuniga A., Dettenhofer M. (2009) Formin1 disruption confers oligodactylism and alters Bmp signaling. Hum. Mol. Genet. 18, 2472–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Quinlan M. E., Hilgert S., Bedrossian A., Mullins R. D., Kerkhoff E. (2007) Regulatory interactions between two actin nucleators, Spire and Cappuccino. J. Cell Biol. 179, 117–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou F., Leder P., Martin S. S. (2006) Formin-1 protein associates with microtubules through a peptide domain encoded by exon-2. Exp. Cell Res. 312, 1119–1126 [DOI] [PubMed] [Google Scholar]
- 15. Vizcarra C. L., Kreutz B., Rodal A. A., Toms A. V., Lu J., Zheng W., Quinlan M. E., Eck M. J. (2011) Structure and function of the interacting domains of Spire and Fmn-family formins. Proc. Natl. Acad. Sci. U.S.A. 108, 11884–11889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Luschnig S., Moussian B., Krauss J., Desjeux I., Perkovic J., Nüsslein-Volhard C. (2004) An F1 genetic screen for maternal-effect mutations affecting embryonic pattern formation in Drosophila melanogaster. Genetics 167, 325–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Singleton K., Woodruff R. I. (1994) The Osmolarity of Adult Drosophila Hemolymph and Its Effect on Oocyte-Nurse Cell Electrical Polarity. Dev. Biol. 161, 154–167 [DOI] [PubMed] [Google Scholar]
- 18. MacLean-Fletcher S., Pollard T. D. (1980) Identification of a factor in conventional muscle actin preparations which inhibits actin filament self-association. Biochem. Biophys. Res. Commun. 96, 18–27 [DOI] [PubMed] [Google Scholar]
- 19. Bor B., Vizcarra C. L., Phillips M. L., Quinlan M. E. (2012) Autoinhibition of the formin Cappuccino in the absence of canonical autoinhibitory domains. Mol. Biol. Cell 23, 3801–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McGhee J. D., von Hippel P. H. (1974) Theoretical aspects of DNA-protein interactions: Co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 86, 469–489 [DOI] [PubMed] [Google Scholar]
- 21. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 22. Hansen S. D., Mullins R. D. (2010) VASP is a processive actin polymerase that requires monomeric actin for barbed end association. J. Cell Biol. 191, 571–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kovar D. R., Harris E. S., Mahaffy R., Higgs H. N., Pollard T. D. (2006) Control of the Assembly of ATP- and ADP-Actin by Formins and Profilin. Cell 124, 423–435 [DOI] [PubMed] [Google Scholar]
- 24. Neidt E. M., Scott B. J., Kovar D. R. (2009) Formin Differentially Utilizes Profilin Isoforms to Rapidly Assemble Actin Filaments. J. Biol. Chem. 284, 673–684 [DOI] [PubMed] [Google Scholar]
- 25. Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M. R., Appel R. D., Bairoch A. (2005) in The Proteomics Protocols Handbook (Walker J. M., ed.), pp. 571–607, Humana Press, Totowa, NJ [Google Scholar]
- 26. Xu Y., Moseley J. B., Sagot I., Poy F., Pellman D., Goode B. L., Eck M. J. (2004) Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell 116, 711–723 [DOI] [PubMed] [Google Scholar]
- 27. Lu J., Meng W., Poy F., Maiti S., Goode B. L., Eck M. J. (2007) Structure of the FH2 Domain of Daam1: Implications for Formin Regulation of Actin Assembly. J. Mol. Biol. 369, 1258–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gould C. J., Maiti S., Michelot A., Graziano B. R., Blanchoin L., Goode B. L. (2011) The Formin DAD Domain Plays Dual Roles in Autoinhibition and Actin Nucleation. Curr. Biol. 21, 384–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dahlgaard K., Raposo A. A., Niccoli T., St Johnston D. (2007) Capu and Spire assemble a cytoplasmic actin mesh that maintains microtubule organization in the Drosophila oocyte. Dev. Cell 13, 539–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Quinlan M. E. (2013) Direct interaction between two actin nucleators is required in Drosophila oogenesis. Development 140, 4417–4425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cheng L., Zhang J., Ahmad S., Rozier L., Yu H., Deng H., Mao Y. (2011) Aurora B Regulates Formin mDia3 in Achieving Metaphase Chromosome Alignment. Dev. Cell 20, 342–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lewkowicz E., Herit F., Le Clainche C., Bourdoncle P., Perez F., Niedergang F. (2008) The microtubule-binding protein CLIP-170 coordinates mDia1 and actin reorganization during CR3-mediated phagocytosis. J. Cell. Biol. 183, 1287–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wen Y., Eng C. H., Schmoranzer J., Cabrera-Poch N., Morris E. J., Chen M., Wallar B. J., Alberts A. S., Gundersen G. G. (2004) EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat. Cell Biol. 6, 820–830 [DOI] [PubMed] [Google Scholar]
- 34. Dehmelt L., Halpain S. (2005) The MAP2/Tau family of microtubule-associated proteins. Genome Biol. 6, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Akhmanova A., Steinmetz M. O. (2008) Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 9, 309–322 [DOI] [PubMed] [Google Scholar]
- 36. Parton R. M., Hamilton R. S., Ball G., Yang L., Cullen C. F., Lu W., Ohkura H., Davis I. (2011) A PAR-1-dependent orientation gradient of dynamic microtubules directs posterior cargo transport in the Drosophila oocyte. J. Cell. Biol. 194, 121–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tanaka T., Kato Y., Matsuda K., Hanyu-Nakamura K., Nakamura A. (2011) Drosophila Mon2 couples Oskar-induced endocytosis with actin remodeling for cortical anchorage of the germ plasm. Development 138, 2523–2532 [DOI] [PubMed] [Google Scholar]
- 38. Kobielak A., Pasolli H. A., Fuchs E. (2004) Mammalian formin-1 participates in adherens junctions and polymerization of linear actin cables. Nat. Cell Biol. 6, 21–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dumont J., Million K., Sunderland K., Rassinier P., Lim H., Leader B., Verlhac M. H. (2007) Formin-2 is required for spindle migration and for the late steps of cytokinesis in mouse oocytes. Dev. Biol. 301, 254–265 [DOI] [PubMed] [Google Scholar]
- 40. Dettenhofer M., Zhou F., Leder P. (2008) Formin 1-isoform IV deficient cells exhibit defects in cell spreading and focal adhesion formation. PLoS One 3, e2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pfender S., Kuznetsov V., Pleiser S., Kerkhoff E., Schuh M. (2011) Spire-type actin nucleators cooperate with Formin-2 to drive asymmetric oocyte division. Curr. Biol. 21, 955–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 43. Oliphant T. (2007). Python for Scientific Computing. Comput. Sci. Eng. 9, 10–20 [Google Scholar]