Fig. 2.
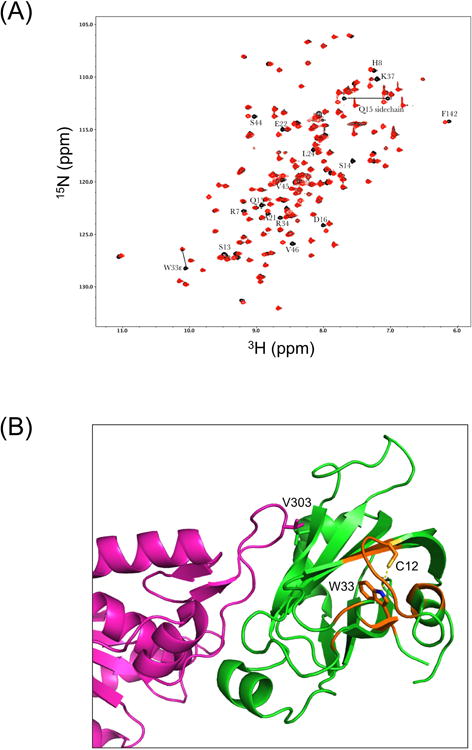
NMR characterization of XRCC1 NTDs. (A) Effect of C12A mutation on the 1H-15N HSQC spectrum of the XRCC1 NTD. The overlayed 1H-15N HSQC spectra for the reduced wild-type (black) and C12A (red) analogs are shown. The W33ε amide shows a large perturbation, consistent with the loss of an H-bonding interaction with the C12 sulfur. Most of these perturbed resonances arise from residues located near C12 and W33, so that changes in the position of W33 will impact the shifts of the amide resonances. In addition to the backbone shifts, a side chain NH2, assigned to Q15 based on its proximity to C12, also exhibits a significant shift perturbation. (B) Perturbed resonances in C12A NTD mapped onto the structure of the reduced wild-type XRCC1 NTD (green):pol β (catalytic domain-magenta) complex, pdb file 3K75. Locations of the perturbed amide resonances are indicated in orange, and the C12 and W33 side chains are also shown.
