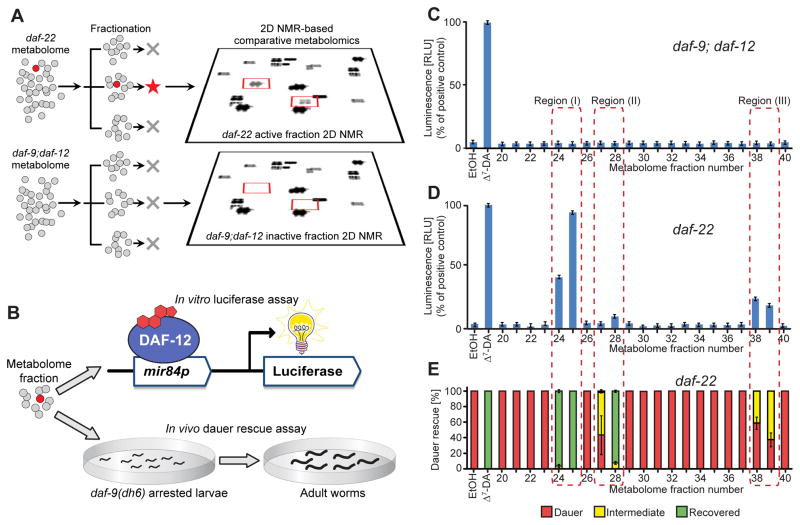
Figure 2. Detection of DAF-12-ligands in C. elegans mutant metabolomes.
(A) Fractionation of active, ligand-rich daf-22 and inactive daf-9;daf-12 metabolomes is followed by 2D NMR-based comparative metabolomics of active fractions.
(B) Assessment of DAF-12-ligand content using (1) an in vitro luciferase assay in HEK-293T cells transfected with full-length DAF-12 and a mir84p-luciferase reporter vector and (2) in vivo daf-9(dh6) dauer rescue assays.
(C) daf-9;daf-12 metabolome fractions are inactive in the luciferase assay. 100 nM Δ7-DA is used as a positive control (error bars, ±SD).
(D) Luciferase assays of daf-22 metabolome fractions reveal three active regions (error bars, ±SD).
(E) daf-9(dh6) dauer rescue assays of daf-22 metabolome fractions show activity in the same three regions (error bars, ±SD). For worm images of scored phenotypes see Figure S1C.
See also Figure S1