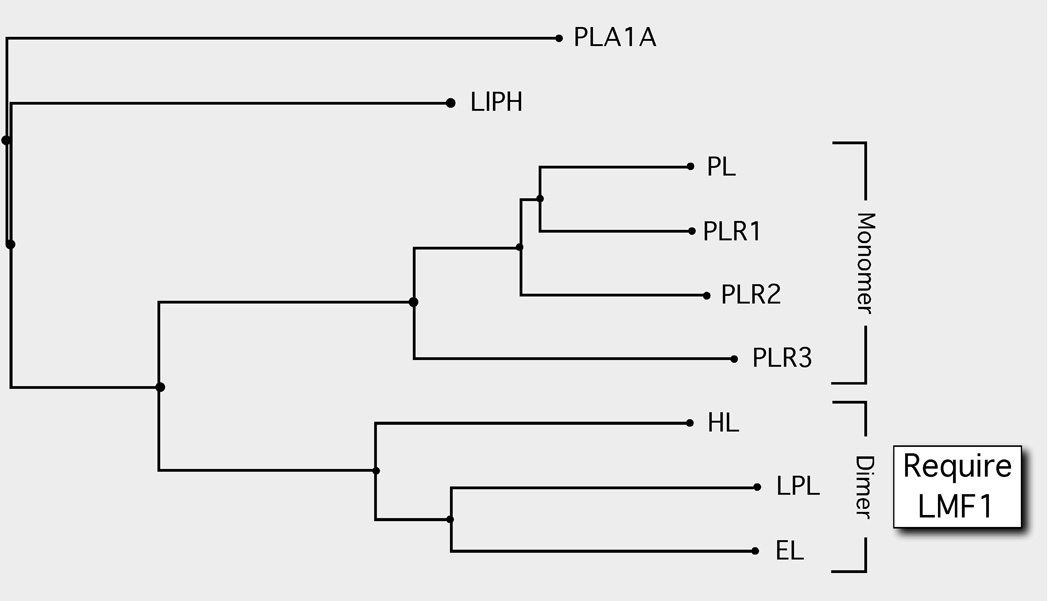
Figure 1. The LMF1-dependent dimeric lipases are a select group within the larger lipase gene family.
Phylogenetic tree of the lipase gene family. Members of the family are secreted enzymes, and thus all mature within the ER. The members include: PLA1A, Phospholipase A1 member A (Q53H76); LIPH, Lipase member H (Q8WWY8); PL, Pancreatic triacylglycerol lipase (P16233) and the three PL-related lipases PLR1 (P54315), PLR2 (P54317) and PLR3 (Q17RR3); HL, Hepatic triacylglycerol lipase (P11150); LPL, Lipoprotein lipase (P06858); and EL, Endothelial lipase (Q9Y5X9). The monomeric state of the PL-related lipases 1, 2 and 3 are inferred based on the parental PL enzyme. The homodimeric state of HL, LPL and EL has been experimentally determined. The subunit structure and requirement for LMF1 is not known for either PLA1A or LIPH. Protein sequence alignments and phylogenetic tree construction was conducted using COBALT [23].