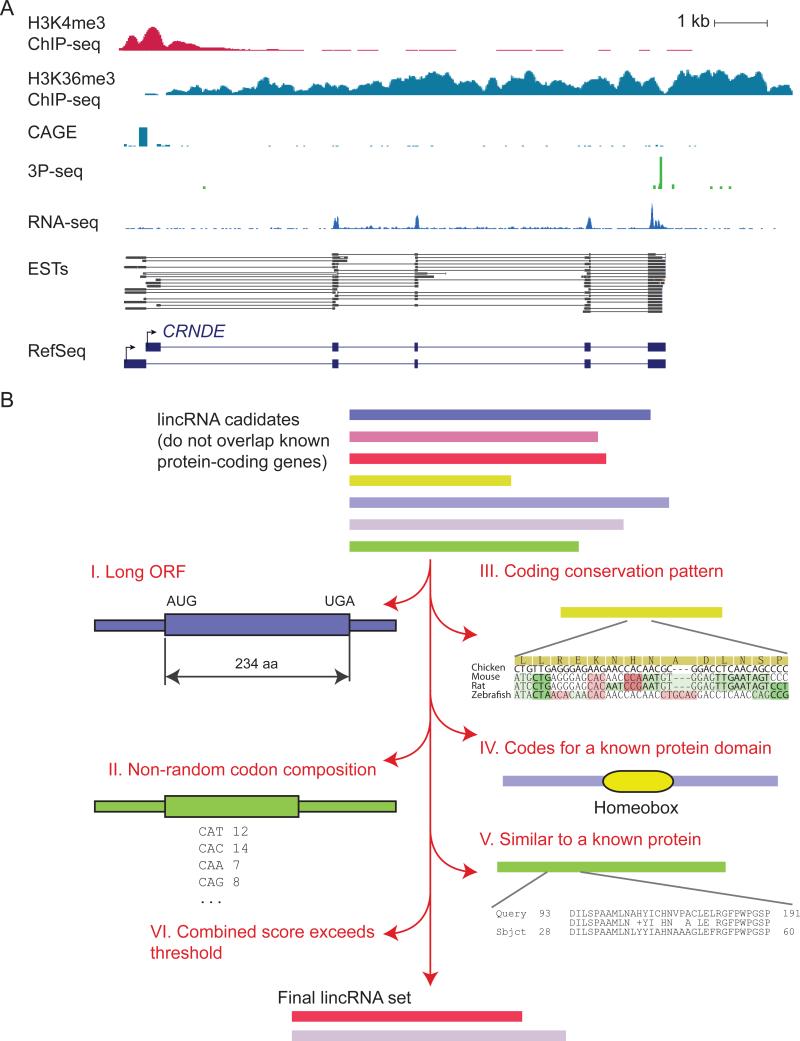
Figure 1. Assembling lincRNA Collections.
(A) Data sets useful for constructing lincRNA transcript models. Information from the indicated genome-wide data sets are plotted for the CRNDE lincRNA locus (chr16:54,950,197-54,963,922 in the human hg19 assembly). A subset of ESTs from GenBank and the corresponding RefSeq annotations are also shown. ChIP-seq and CAGE (ENCODE project, HeLaS3 cells), 3P-Seq (HeLa cells, C. Jan and D.P.B., unpublished data), RNA-seq (HeLa cells; Guo et al., 2010) were plotted using the UCSC genome browser.
(B) A generic lincRNA annotation pipeline, illustrating criteria used to filter potential mRNAs from the list of candidates.