Abstract
Celiac disease (CD) is an immune mediated, polygenic disorder, where HLA-DQ2/DQ8 alleles contribute around 35% to genetic risk, but several other genes are also involved. Genome-wide association studies (GWASs) and the more recent immunochip genotyping projects have fine-mapped 39 regions of genetic susceptibility to the disease, most of which harbor candidate genes that could participate in this disease process. We focused our attention to the GWAS peak on chr6: 127.99–128.38 Mb, a region including two genes, thymocyte-expressed molecule involved in selection (THEMIS) and protein tyrosine phosphatase, receptor type, kappa (PTPRK), both of which have immune-related functions. The aim of this work was to evaluate the expression levels of these two genes in duodenal mucosa of active and treated CD patients and in controls, and to determine whether SNPs (rs802734, rs55743914, rs72975916, rs10484718 and rs9491896) associated with CD have any influence on gene expression. THEMIS showed higher expression in active CD compared with treated patients and controls, whereas PTPRK showed lower expression. Our study confirmed the association of this region with CD in our population, but only the genotype of rs802734 showed some influence in the expression of THEMIS. On the other hand, we found a significant positive correlation between THEMIS and PTPRK mRNA levels in CD patients but not in controls. Our results suggest a possible role for both candidate genes in CD pathogenesis and the existence of complex, regulatory relationships that reside in the vast non-coding, functional intergenic regions of the genome. Further investigation is needed to clarify the impact of the disease-associated SNPs on gene function.
Keywords: celiac disease, genetic association, THEMIS, PTPRK, gene expression
Introduction
Celiac disease (CD) is a common (1:100) immune-mediated enteropathy that develops in genetically susceptible individuals by ingestion of gluten and related proteins from wheat, barley, rye and probably oats. Villous atrophy, crypt hyperplasia and lymphocytic infiltration of the small intestinal mucosa are the typical histological findings in active CD, although milder changes in the mucosa are increasingly common at diagnosis. Clinical presentation varies from typical gastrointestinal symptoms to extraintestinal manifestations. The only effective treatment consists in a lifelong gluten-free diet (GFD).1 CD is a multifactorial, polygenic disorder, and the most important genetic susceptibility determinant is located in the MHC locus; HLA-DQ2 (DQA1*05, DQB1*02) and/or HLA-DQ8 (DQA1*03, DQB1*03:02) are necessary, although not sufficient, for disease development. However, these susceptibility alleles only explain 35% of genetic risk and several other genes also contribute to CD pathogenesis.2
To date, the two genome-wide association studies (GWAS) performed in CD have revealed 26 regions of genetic susceptibility to the disease.2, 3 Several genes within those regions have been proposed as etiological candidates, most of them related to immune response pathways (RGS1, IL18R1/IL18RAP, CCR3, IL12A/SCHIP1, TAGAP, SH2B3, BACH2, CCR4, CD80, CIITA-SOCS1-CLEC16A, ICOSLG, ZMIZ1) or to T-cell maturation (ETS1, RUNX3, THEMIS, PTPRK or TNFRSF14) and it has been proposed that they might participate in different steps in the pathogenesis of CD. More recently, the immunochip genotyping array has fine-mapped these regions and reported additional 13 susceptibility loci.4 Many of the 39 non-HLA regions associated with the genetic risk to CD development are also shared with other autoimmune diseases; in particular, type 1 diabetes and rheumatoid arthritis.
In this study, we focused our attention in the GWAS peak on the chr6: 127.99–128.38 Mb HapMap LD block, which includes two genes: thymocyte-expressed molecule involved in selection (THEMIS) and protein tyrosine phosphatase, receptor type, kappa (PTPRK), both of which have immune-related functions, and are thus potential candidate genes in CD pathogenesis. THEMIS expression is limited to lymphoid tissues, and studies in mice have shown that it encodes an important molecule for T-cell development, being expressed at higher levels in pre-TcR thymocytes.5 Although its expression is reduced in differentiated T lymphocytes,6 it is thought to have a role in TcR activation signaling.7 In turn, PTPRK performs functions related to the maintenance of cell junctions8, 9, 10 and in the modulation of epidermal growth factor receptor (EGFR) activity, resulting in an inhibition of cell proliferation.11, 12 Interestingly, both THEMIS and PTPRK have been implicated in normal CD4+ T-cell development in rodents, suggesting that they participate in common biological processes.13 The GWAS peak in this region was marked by SNP rs802734 (11 kb 3′ of PTPRK and 39 kb 5′ from THEMIS). The fine-mapping immunochip study revealed a major association peak spanning 2 kb (chr6: 128 332–128 335 kb) marked by rs55743914 (in the 3′-UTR of PTPRK) and a broader secondary signal (chr6: 128 307–128 339 kb) marked by rs72975916, tipping the scale in favor of PTPRK.4 However, no functional confirmation of these candidate genes has been performed.
The objective of this work was to evaluate the expression levels of both THEMIS and PTPRK in duodenal mucosa of active and treated CD patients and controls, as well as the effect of in vitro gliadin challenge on biopsies from CD patients on GFD. We also aimed to determine whether disease-associated variants have any influence on gene expression, considering genotypes of the top SNPs in GWAS and the immunochip projects (rs802734, rs55743914 and rs72975916) as well as those that were most strongly associated in our population (rs10484718 and rs9402010).
Materials and methods
Patients and samples
CD patients were diagnosed at the Pediatric Gastroenterology Unit of Cruces University Hospital, according to the European Society of Pediatric Gastroenterology, Hepatology, and Nutrition (ESPGHAN) criteria. For gene expression analyses, distal duodenal biopsy pairs from 36 pediatric CD patients (22 females; age at diagnosis 2.48±1.19 years) were taken at the time of diagnosis (on a gluten-containing diet, with CD-associated antibodies and atrophy of intestinal villi and crypt hyperplasia) and at a routine visit during disease remission, after at least 2 years on GFD (asymptomatic, negative serology for autoantibodies against anti-transglutaminase 2 and normalized histology of the intestinal epithelium). Twelve tissue samples from non-celiac individuals were used as controls. For in vitro stimulation studies, two portions of duodenal specimens taken from eight CD patients in remission (as above) were incubated immediately in RPMI, alone or with 250 μg/ml pepsin–trypsin digested gliadin, at 37 °C and 5% CO2 during 4 h. Tissue samples were conserved in liquid nitrogen until RNA extraction. DNA from the same biopsy donors was obtained from peripheral blood using NucleoSpin Blood Kit (Machery Nagel GmbH & Co. KG, Düren, Germany). The study was approved by the Cruces University Hospital Ethics Board and samples were collected after informed consent had been obtained from the subjects and/or their parents.
Peptic tryptic digestion of gliadin
Gliadin (500 mg) from wheat (Sigma-Aldrich Co., St Louis, MO, USA, cat no. G3375) was dissolved in 5 ml 0.2 ℳ HCl and incubated with 5 mg pepsin (Sigma-Aldrich Co., cat no. P6887) with continuous stirring at 37 °C for 2 h. At the end of this incubation, pH was adjusted to 7.4 with NaOH and the mixture was incubated with 5 mg trypsin (Sigma-Aldrich Co., cat no. T9201) for 4 h at 37 °C. To inactivate the enzymes, the solution was heated in a boiling water bath for 30 min. Finally, the digest was centrifuged at 2000 g for 10 min, and the supernatant was sterilized by filtering through a 20 μm pore membrane. Protein content was quantified by BCA method.
Gene expression analyses
Frozen biopsies were disrupted with disposable plastic pestles in 300 μl lysis buffer and homogenized in QIAshredder columns (Qiagen GmbH, Hilden, Germany). Total RNA was isolated using an RNeasy Micro Kit (Qiagen) with DNase I treatment, and stored at −80 °C until use. The expression of THEMIS and PTPRK was quantified by real-time, reverse transcriptase PCR (RT-PCR) using commercial Assay-on-Demand primer-probe sets (Applied Biosystems, Foster City, CA, USA) and a single reaction enzyme mixture (QuantiTect Probe RT-PCR kit, Qiagen) with 8 ng of total RNA. Experiments were performed in triplicate in an ABI7900HT system and housekeeping gene RPLPO was simultaneously quantified and used as an endogenous control of input RNA. Relative expression in each sample was calculated using the accurate Ct method and normalized to one of the control samples, as previously described.14
Association analysis, SNP selection and genotyping assays
To identify association signals specific to our population, the immunochip genotyping results from the CEGEC (Spanish Consortium for the Genetics of Celiac Disease) sample set (545 CD patients and 308 controls) were analyzed independently using PLINK v.1.07 (http://pngu.mgh.harvard.edu/purcell/plink).15 The top GWAS and immunochip SNPs (overall and specific for the CEGEC cohort) were genotyped in biopsy donors using commercially available TaqMan allelic discrimination assays (rs802734, rs72975916, rs10484718 and rs9491896, a proxy for rs9402010) or a custom assay designed using Primer Express Software for rs55743914 (forward primer: 5′-ACCCACAGATATCTCTCGTTAGAAGTAA-3′, reverse primer: 5′-GGCTCGTAAAGTCAGAACATAGCTT-3′, probes: 5′-FAM-TCTGCATACAGTTTCACCTA-MGB-3′ and 5′-VIC-CATACAGCTTCACCTAGA-MGB-3′) in an 7900HT Real-Time PCR sequence detection system (all from Applied Biosystems). The putative effects of the genotyped SNPs on predicted splicing sites and microRNA or transcription factor-binding sites were analyzed with the online tool PupaSuite (http://pupasuite.bioinfo.cipf.es)16 and in the Haploreg web site (http://www.broadinstitute.org/mammals/haploreg/haploreg.php).17
Statistic tests and correlation analyses
Gene expression levels were compared between CD patients at diagnosis and after treatment using paired T-tests. Comparison of the expression levels between control subjects and both active and treated CD patients, as well as among SNP genotype patient groups were performed using unpaired T-tests. The relationship between the levels of expression of each gene in active disease and SNP genotype was calculated using Pearson's correlation test. All analyses were performed with Prism v.5.0 software (GraphPad software Inc., La Jolla, CA, USA) and two-tailed P-values below 0.05 were considered significant.
Results and Discussion
Association studies have proven useful for the search of complex disease susceptibility regions, but positive signals have to be replicated in different populations in order to confirm preliminary results. Moreover, it is also important to perform functional studies in the disease target tissue to detect putative effects of the associated polymorphisms on the genes surrounding those variants, and thus identify the causative mechanism underlying the association. In this study, we have focused on the CD association peak on chr6: 127.99–128.38 Mb, a region that harbors THEMIS and PTPRK genes, and have performed genetic and functional analyses in CD patients and controls.
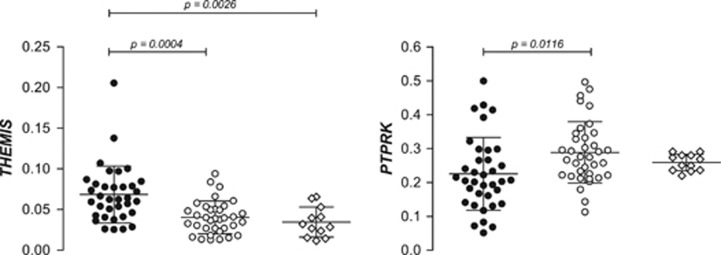
Gene expression analysis of duodenal biopsies from active CD patients showed higher levels of THEMIS mRNA compared with samples taken from the same patients after GFD treatment or from controls (Figure 1). This finding is coherent with the disease picture, as THEMIS is a lymphoid cell-specific gene and there is a notable inflammatory infiltration in the intestinal mucosa of active CD patients. On the contrary, PTPRK was observed to be underexpressed in active CD intestinal mucosa compared with treated individuals, and a similar trend was seen compared with non-celiac individuals, although P-values did not reach statistical significance (Figure 1). Given its role in the maintenance of cell–cell junctions, the low levels of PTPRK in active disease are in accordance with the increased barrier permeability observed in active CD, and its inhibition could also favor the hyperproliferation of intestinal crypt cells. On the other hand, treated CD patients showed similar expression levels compared with non-celiac controls, indicating that the adherence to GFD is sufficient to return the transcriptional activity of both genes to normal levels. The latter would indicate that the changes in expression are likely to be a consequence of the chronic mucosal inflammation typical of active CD, rather than a genetically determined baseline alteration driving a functional predisposition to CD.
Figure 1.
Relative expression of THEMIS and PTPRK in intestinal mucosa from active (black circles) and treated (white circles) CD patients and non-celiac controls (white diamonds). P-values correspond to a paired t-test in the case of active vs treated CD comparisons, and to an unpaired t-test when comparing patients with controls.
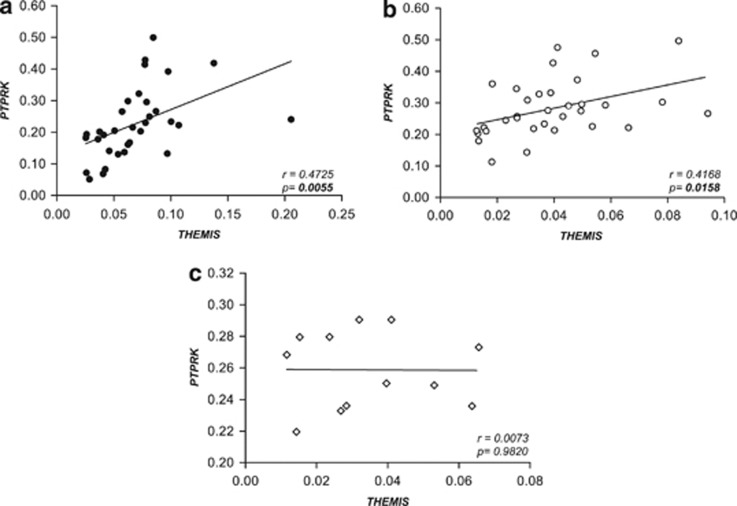
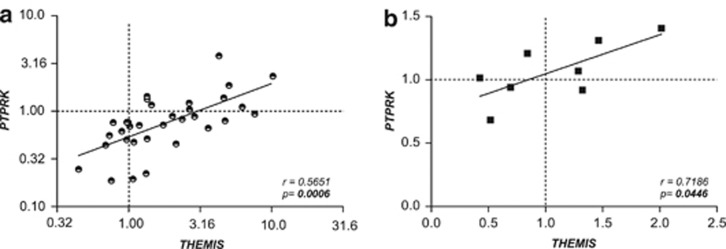
Interestingly, there was a robust correlation between the expression of both genes in active and treated CD that was absent in the non-celiac control group (Figure 2). Analyses comparing fold change values induced by disease activation (active CD/treated CD gene expression levels) revealed that the changes observed in both genes followed the same trend in each individual sample (Figure 3a). No additional non-coding transcripts that could have trans effects elsewhere in the genome have been detected in this region, so it seems plausible that these two genes could share some cis-acting regulatory element specifically related to the disease. Indeed, according to the recently released ENCODE data, many of the SNPs genotyped in this region (and others in LD) fall into putative genomic regulatory elements (protein-binding motifs, histone marks, DNAse hypersensitivy, and so on—Supplementary Table 1). Moreover, in vitro stimulation of biopsy samples with pepsin–trypsin digested gliadin further supports this common regulation hypothesis. Neither THEMIS nor PTPRK expression was significantly modulated by the gliadin insult under the incubation conditions used, but once more, gliadin-induced fold change values of both genes showed positive correlation (Figure 3b).
Figure 2.
Correlation between THEMIS and PTPRK expression in intestinal mucosa from active (a, black circles) and treated (b, white circles) CD patients and non-celiac controls (c, white diamonds). Pearson's correlation coefficient r and P-values of the linear regression analyses are shown.
Figure 3.
Correlation of expression fold changes for PTPRK and THEMIS (a) in intestinal mucosa from CD patients taken at diagnosis (active) compared with after >2 years on GFD (treated) and (b) induced by in vitro gliadin stimulation of intestinal biopsy samples from treated CD patients. Pearson's correlation coefficient r and P-values of the linear regression analyses are shown. In (a), logarithmic values are represented, but the axes correspond to the linear values.
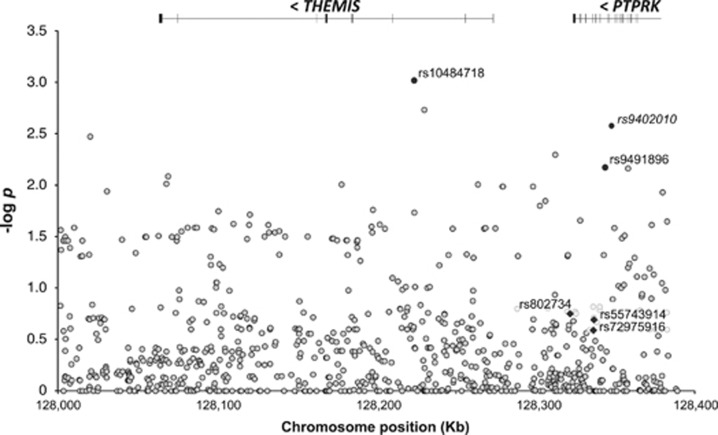
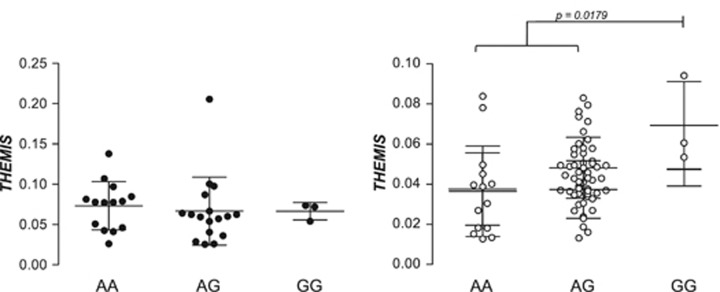
The analysis of the immunochip data from this region in CEGEC samples did not show replication of the GWAS (rs802734) or immunochip (rs55743914 and rs72975916) top-associated SNPs, although several other SNPs in the region showed nominal association in our population (Figure 4). The three overall top SNPs and the two variants with the lowest P-value in our cohort (rs10484718 and rs9491896, a proxy for rs9402010) were genotyped in tissue sample donors to evaluate whether they could influence THEMIS or PTPRK gene expression levels. None of the SNPs had any effect on PTPRK expression, but THEMIS mRNA levels were significantly higher in treated patients homozygous for the risk-associated rs802734*G allele, identified in the initial GWAS (Figure 5). However, these results must be interpreted with caution, given the low frequency of the homozygous GG genotype among patients and the fact that the strong association with this SNP was not confirmed in the larger immunochip cohort. Besides, no functional implication of rs802734 appears in the ENCODE data.
Figure 4.
Association analysis results in the CEGEC population for the immunochip SNPs in the PTPRK–THEMIS genomic region (chr6: 128.0–128.4 kb). SNP distribution, location of the genes and log10 of nominal P-values of an allelic test are shown. SNPs that were analyzed in tissue sample donors include variants that had shown the strongest association with CD in the GWAS3 and the overall immunochip project4 (black diamonds) and top SNPs from this study (black circles), except for rs9402010, for which a Taqman allelic discrimination assay could not be designed and was replaced by rs9491896.
Figure 5.
Influence of rs802734 genotype on THEMIS expression in duodenal mucosa from active (black circles) and GFD-treated (white circles) CD patients. The genotypes of rs55743914, rs72975916, rs10484718 and rs9491896 did not have any effect on THEMIS or PTPRK expression (data not shown).
The identification of the functional effects of association signals in the risk to complex diseases remains a challenging task that is far from being straightforward.18 None of the SNPs studied has demonstrated to have a clear, direct influence on the expression of neighboring genes that could link association with causality. However, our study suggests the existence of complex, regulatory relationships that reside in the vast non-coding, functional genome. Although changes in expression levels seem to be a consequence of the inflammation present in untreated CD, the correlation observed between the two genes is present both at diagnosis and on tissue normalization by GFD, as well as after a short in vitro gliadin challenge, suggesting a common regulatory mechanism that could be due to functional genomic variants within this region. Further investigation is needed to clarify the role of THEMIS and PTPRK in the pathogenesis of CD and to identify the mechanisms by which they are corregulated in the CD gut, and thus contribute to elucidate the molecular mechanisms by which associated genetic variants influence the risk to complex diseases.
Acknowledgments
This work was partially funded by Research Project grants from the Instituto de Salud Carlos III of the Spanish Ministry of Economy and Competitiveness (PI10/0310) and from the Basque Department of Industry (SAIO-PE08BF03) to JRB. CB is a predoctoral fellow of the CONICET Program (Argentina) and also received a fellowship for a short visit to JRB's laboratory (Beca de Formación Permanente from Fundación Carolina, Spain). FC is member of the Research Career of CONICET Program (Argentina). NF-J and LP-I are predoctoral fellows supported by FPI grants from the Basque Department of Education, Universities and Research (BIF-2009-099 and BIF-2010-189, respectively). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
APPENDIX
CEGEC (Spanish Consortium for the Genetics of Celiac Disease) Participants
Montserrat Alsina, María Esteve and Laura Pardo (CATLAB-Hospital Universitari Mutua de Terrassa-Barcelona); Luis Castaño, Galder Gutierrez-Ziardegi and Juan Carlos Vitoria (Hospital Universitario de Cruces, Barakaldo-Bizkaia); Luis Rodrigo, Carlos López-Larrea and Antonio López-Vázquez (Hospital General de Asturias, Oviedo-Asturias); Idoia Hualde and Zuriñe García (Hospital de Txagorritxu, Vitoria-Gasteiz); Blanca Hernández, María Antonia Ramos-Arroyo and Félix Sánchez-Valverde (Hospital Virgen del Camino, Pamplona-Iruña); Carme Farré, Teresa Marqués and Pere Vilar (Hospital Sant Joan de Deu, Esplugues de Llobregat-Barcelona), Eduardo Arranz and José Antonio Garrote (Universidad de Valladolid, Valladolid).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Abadie V, Sollid LM, Barreiro LB, Jabri B. Integration of genetic and immunological insights into a model of celiac disease pathogenesis. Annu Rev Immunol. 2011;29:493–525. doi: 10.1146/annurev-immunol-040210-092915. [DOI] [PubMed] [Google Scholar]
- Dubois PC, Trynka G, Franke L, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel DA, Franke L, Hunt KA, et al. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat Genet. 2007;39:827–829. doi: 10.1038/ng2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trynka G, Hunt KA, Bockett NA, et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet. 2011;43:1193–1201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesourne R, Uehara S, Lee J, et al. Themis, a new T cell specific protein important for late thymocyte development. Nat Immunol. 2009;10:840–847. doi: 10.1038/ni.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu G, Vallée S, Rybakin V, et al. Themis controls thymocyte selection through modulation of T cell antigen receptor-mediated signalling. Nat Immunol. 2009;10:848–856. doi: 10.1038/ni.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmeyer C, Paster W, Pepper D, et al. T cell receptor (TCR)-induced tyrosine phosphorylation dynamics identifies THEMIS as a new TCR signalosome component. J Biol Chem. 2011;286:7535–7547. doi: 10.1074/jbc.M110.201236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sap J, Jiang YP, Friedlander D, Grumet M, Schlessinger J. Receptor tyrosine phosphatase R-PTP-kappa mediates homophilic binding. Mol Cell Biol. 1994;14:1–9. doi: 10.1128/mcb.14.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnekenburger J, Mayerle J, Krüger B, et al. Protein tyrosine phosphatase kappa and SHP-1 are involved in the regulation of cell-cell contacts at adherens junctions in the exocrine pancreas. Gut. 2005;54:1445–1455. doi: 10.1136/gut.2004.063164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders L, Mertins P, Lammich S, et al. Furin-, ADAM 10-, and gamma-secretase-mediated cleavage of a receptor tyrosine phosphatase and regulation of beta-catenin's transcriptional activity. Mol Cell Biol. 2006;26:3917–3934. doi: 10.1128/MCB.26.10.3917-3934.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Shao Y, Voorhees JJ, Fisher GJ. Oxidative inhibition of receptor type protein tyrosine phosphatase kappa by ultraviolet irradiation activates epidermal growth factor receptor in human keratinocytes. J Biol Chem. 2006;281:27389–27397. doi: 10.1074/jbc.M602355200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Baker D, Quan T, Baldassare JJ, Voorhees JJ, Fisher GJ. Receptor type protein tyrosine phosphatase-kappa mediates cross-talk between transforming growth factor-beta and epidermal growth factor receptor signaling pathways in human keratinocytes. Mol Biol Cell. 2010;21:29–35. doi: 10.1091/mbc.E09-08-0710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata R, Sasaki N, Agui T. Contiguous gene deletion of Ptprk and Themis causes T-helper immunodeficiency (thid) in the LEC rat. Biomed Res. 2010;31:83–87. doi: 10.2220/biomedres.31.83. [DOI] [PubMed] [Google Scholar]
- Martin-Pagola A, Perez-Nanclares G, Ortiz L, et al. MICA response to gliadin in intestinal mucosa from celiac patients. Immunogenetics. 2004;56:549–554. doi: 10.1007/s00251-004-0724-8. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde L, Vaquerizas JM, Dopazo H, et al. PupaSuite: finding functional SNPs for large-scale genotyping purposes. Nucleic Acid Res. 2006;34:W621–W625. doi: 10.1093/nar/gkl071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WardL KellisM. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acid Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaza-Izurieta L, Castellanos-Rubio A, Irastorza I, et al. Revisiting genome wide association studies (GWAS) in coeliac disease: replication study in Spanish population and expression analysis of candidate genes. J Med Genet. 2011;48:493–496. doi: 10.1136/jmg.2011.089714. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.