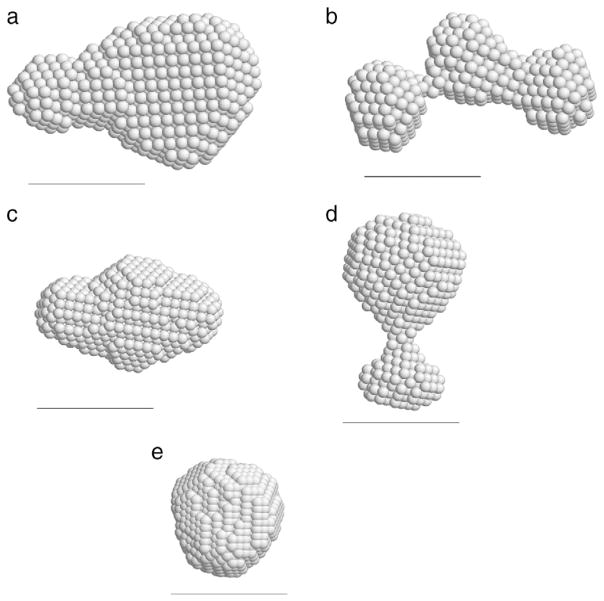
Fig. 4.
Calculated structures by DAMMIF [28] and DAMAVER [29] programs. The scale bar is 30 Å. Structures (a), (b) and (d) were calculated from experimental data and structures (c) and (e) were calculated from atomic models: scattering profiles were obtained from PDB coordinates and then DAMMIF calculations were done with this scattering curve. (a) hNck2 SH3 domain at pH 8 (3.3 mg/ml). Rmax was set at 70 Å. (b) hNck2 SH3 domain at pH 2 (3.2 mg/ml). Rmax was set at 65 Å. (c) dimer of Esp8 SH3 domain at pH 7. PDB code is 1I07 [41], (64 × 2 residues). (d) hNck2 SH3 domain at pH 6 (0.14 mg/ml). Rmax was set at 55 Å. (e) hNck2 SH3 domain. PDB code is 2B86 (59 residues) [27].