Abstract
To maintain growth and division, cells require a large-scale production of rRNAs which occurs in the nucleolus. Recently, we have shown the interaction of nucleolar phosphatidylinositol 4,5-bisphosphate (PIP2) with proteins involved in rRNA transcription and processing, namely RNA polymerase I (Pol I), UBF, and fibrillarin. Here we extend the study by investigating transcription-related localization of PIP2 in regards to transcription and processing complexes of Pol I. To achieve this, we used either physiological inhibition of transcription during mitosis or inhibition by treatment the cells with actinomycin D (AMD) or 5,6-dichloro-1β-d-ribofuranosyl-benzimidazole (DRB). We show that PIP2 is associated with Pol I subunits and UBF in a transcription-independent manner. On the other hand, PIP2/fibrillarin colocalization is dependent on the production of rRNA. These results indicate that PIP2 is required not only during rRNA production and biogenesis, as we have shown before, but also plays a structural role as an anchor for the Pol I pre-initiation complex during the cell cycle. We suggest that throughout mitosis, PIP2 together with UBF is involved in forming and maintaining the core platform of the rDNA helix structure. Thus we introduce PIP2 as a novel component of the NOR complex, which is further engaged in the renewed rRNA synthesis upon exit from mitosis.
Keywords: PIP2, mitosis, transcription, nucleolus, RNA polymerase I, UBF, fibrillarin
Introduction
The nucleolus is a prominent structure within the cell nucleus which forms around the nucleolar organizing regions (NORs) in a cell cycle-dependent manner. The nucleolus is the cell ribosome factory and, in addition, it is also a multifunctional domain involved in a variety of processes and severe diseases including stress response, biogenesis of ribonucleoproteins, ribosomopathies and cancer (for reviews see refs. 1 and 2). It is therefore important to understand the function and regulation of this crucial cell compartment.
Throughout interphase, cells produce ribosomes with the maximal yield in G2 phase. The sequential stages of ribosome biogenesis are reflected in the general architecture of the nucleolus comprising three main sub-compartments, which have been well defined by electron microscopy: fibrillar centers (FCs), dense fibrillar component (DFC), and granular component (GC). Transcription of ribosomal DNA (rDNA) by RNA polymerase I (Pol I) takes place mostly at the FC/DFC border. Pol I is a multi-polypeptide complex composed of constant subunits and temporarily associated factors. The main component of the Pol I machinery is the upstream binding factor (UBF). This architectural protein comprises six high mobility group (HMG) boxes enabling a single dimer of UBF to induce an almost 360° looping in 140 base-pairs of rDNA via multiple co-phased turns, forming the nucleo-protein structure referred to as the rRNA gene enhancesome.3-5 UBF binding to enhancer region of rDNA leads to the creation of open chromatin structure by displacing linker histone H1 and the assembly of pre-initiation complex (PIC) on the promoter.6 PIC formation involves concerted action of UBF and the promoter selectivity factor (SL1) consisting of TATA-binding protein (TBP) and Pol I-specific TBP-associated factors TAFI110, TAFI63, and TAFI48.7,8 UBF recruits SL1 via the interaction with TAFI48 and TBP, where TAFIs provide highly sequence-specific promoter recognition.9-11 UBF also binds extensively across the transcribed regions of the rDNA and maintains architecture of transcriptionally active sites of nucleoli.12 Due to its ability to form rDNA loops, UBF brings together the start site proximal core promoter and the upstream control element providing the correct scaffolding for productive interactions between UBF and SL1.3,13 Thereby, UBF in a complex with SL1 creates a core-helix DNA structure to achieve efficient ribosomal RNA (rRNA) production in a limited nucleolar space.14 Before ribosomal subunits assemble, the rRNA transcripts proceed through several stages of maturation, which take place sequentially in the DFC and GC.15 Fibrillarin is a rRNA 2'-O-methyltransferase that localizes to the DFC region and is involved in the early stages of rRNA processing.16
Upon entry into mitosis, the ribosome production stops in prophase and the nucleolus is then disassembled in a sequential manner. The repression of Pol I transcription is connected with the ordered release of the processing complexes from rDNA transcription machinery. At the end of prophase, upon chromosome condensation and breakdown of nuclear envelope, the interphase nucleolar architecture is not detectable, but the components of rDNA transcription machinery and pre-rRNA processing machinery remain partially assembled and are kept at different sites throughout mitosis. rDNA, that was actively transcribed in the preceding interphase, remains associated with the Pol I subunits, UBF, SL1 subunits and transcription termination factor TTF1 and resides in the mitotic NORs.17-19 The proteins and small nucleolar RNA (snoRNAs) of the processing complexes, ribosomal proteins as well as the pre-rRNA molecules remain assembled and stored in either the perichromosomal compartment or in cytoplasmic nucleolar-derived foci.20,21 At the end of mitosis, the re-assembling of the nucleolus is coordinated via activation of Pol I transcription machinery and rRNA processing machinery. During transferring to the sites of activated rDNA transcription, rRNA processing complexes are temporarily assembled into prenucleolar bodies.20
Phosphatidylinositol 4,5-bisphosphate (PIP2) is a minor lipid found on cellular membranes that binds and regulates activities of numerous proteins. PIP2 is cleaved by phospholipase C (PLC) to inositol 1,4,5-trisphosphate (IP3) and diacylglycerol (DAG). IP3 is released into the cytoplasm resulting in Ca2+ mobilization, and DAG remains bound to the membrane where it activates protein kinase C leading to a cellular response (for a review see ref. 22). Phospholipids and PIP2 in particular have been also shown to be present in the nucleus, where they regulate some of the nuclear functions in nuclear speckles.23-26 We have recently shown that PIP2 binds to the transcription factor UBF and makes a complex with Pol I on the promoter of ribosomal genes. Moreover, depletion of PIP2 results in reduction of Pol I transcription. PIP2 binding to fibrillarin and its colocalization with nascent transcripts in the nucleolus indicates that PIP2 might be involved in both transcription and early processing of rRNA.27
Here we extend this study by investigating transcription-related localization of PIP2 in regards to Pol I transcription and processing complexes. To do this, we have used either native inhibition of transcription during mitosis or inhibition by treatment the cells with different compounds. Actinomycin D (AMD) is known to inhibit Pol I transcription at low doses by binding DNA at the transcription initiation complex and preventing elongation of nascent transcripts. This results in redistribution of nucleolar components and the formation of nucleolar caps and intranuclear inclusions.28 5,6-dichloro-1β-d-ribofuranosyl-benzimidazole (DRB) is a nucleoside analog which inhibits transcript elongation by Pol II. This leads also to decreased levels of important nucleolar proteins and produces a dramatic change in nucleolar architecture by forming bead-like structures, although Pol I transcription is preserved.29 We show that PIP2 is associated with Pol I subunits and UBF in a transcription-independent manner. On the other hand, PIP2/fibrillarin colocalization is dependent on the production of rRNA. These results indicate that PIP2 is required not only during rRNA production and biogenesis, as we have shown before, but also plays a structural role as an anchor for the Pol I pre-initiation complex during the cell cycle. We suggest that throughout mitosis PIP2 together with UBF is involved in forming and maintaining the core platform of the rDNA helix structure. Thus we introduce PIP2 as a novel component of the NOR complex, which is further engaged in the renewed rRNA synthesis upon exit from mitosis.
Results
PIP2 association with Pol I complex is transcription-independent
We showed recently that PIP2 participates in formation of Pol I transcription foci in interphase nucleoli.27 Nevertheless, the dependence of PIP2 localization to the sites of Pol I transcription factory on the transcriptional status of the cell is not known. We chose mitotic cells with physiologically blocked transcription and used an immunoprecipitation assay to detect the components of Pol I transcription machinery still associated with PIP2. We confirmed the detection reliability by monitoring NOR integrity with immunofluorescent microscopy taking the samples during cell fractionation. We specifically detected UBF and two subunits of Pol I, PAF53 and RPA116, in the same complex with PIP2 (Fig. 1).
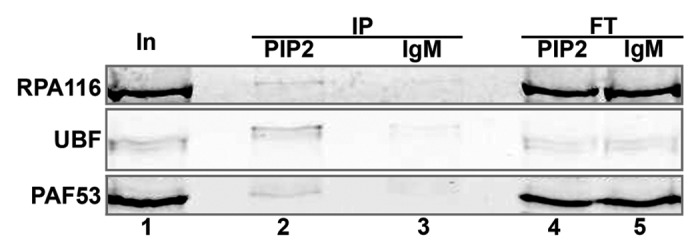
Figure 1. PIP2 binds to the proteins of Pol I pre-initiation complex in mitotic cells. Immunoprecipitation assay of mitotic cell extract using anti-PIP2 antibody and protein L-agarose beads detected UBF and the Pol I subunits, PAF53 and RPA116, in the PIP2-bound protein complex. Lane 1, input; lane 2, protein immunoprecipitated with anti-PIP2 antibody; lane 3, protein immunoprecipitated with isotype control; lane 4, protein unbound to anti-PIP2 antibody (flow-through); lane 5, protein unbound to isotype control (flow-through); In, input; IP, immunoprecipitate; FT, flow-through.
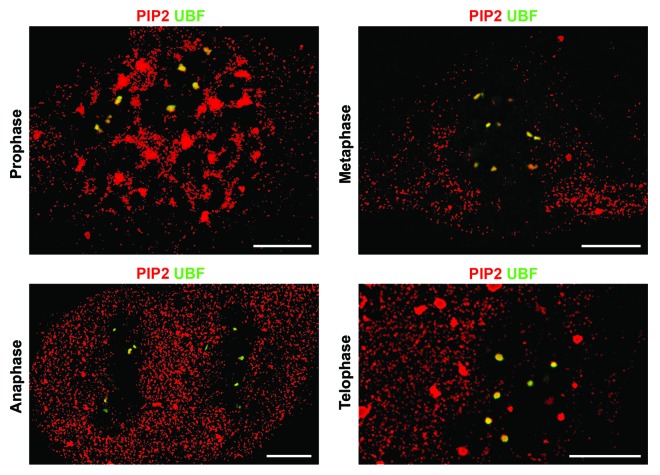
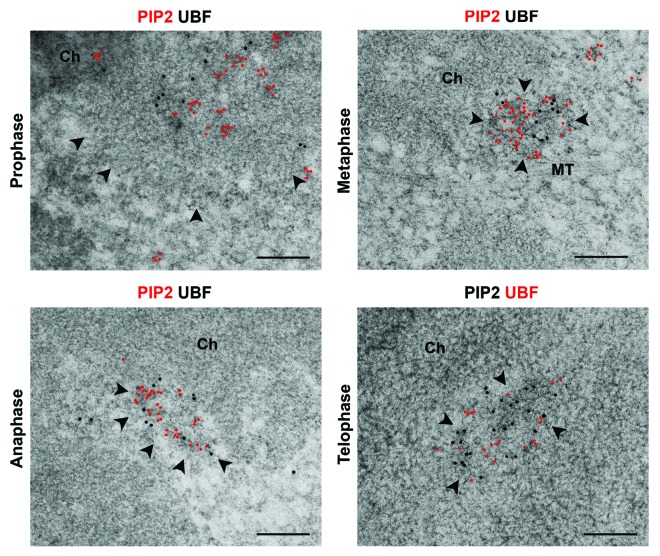
Based on the association of the components of Pol I pre-initiation complex with PIP2 in mitotic cells, we asked whether this association is retained in NORs during all mitotic stages. To test this, we used super-resolution structured illumination microscopy (SIM) combined with multi-immunolabeling approach as well as high-pressure freezing and freeze-substitution (HPF/FS) combined with immunoelectron microscopy (IEM). We showed previously that HPF/FS preserves the near-native state of nuclear compartments and molecular complexes.30 We observed regular colocalization patterns of PIP2 and UBF during different mitotic phases in which transcription is physiologically inhibited. UBF serves as a marker of mitotic NORs due to its persistent presence on extended fibers of rDNA during the entire cell cycle.31 SIM showed colocalization between PIP2 and UBF in NORs throughout prophase and metaphase, continuing in “daughter” NORs in anaphase and telophase (Fig. 2). HPF/FS IEM confirmed this colocalization of PIP2 with UBF in clearly distinguishable NORs, characterized by less compacted structure as compared with the bulk of condensed chromatin, from prophase to telophase (Fig. 3).
Figure 2. PIP2 retains its colocalization with UBF during mitosis as demonstrated by SIM. Multi-immunolabeling followed by super-resolution structured illumination microscopy showed that PIP2 colocalizes with UBF in NORs, which are the mitotic counterparts of the interphase FCs, in prophase and metaphase. This colocalization pattern persists in anaphase and telophase in “daughter” NORs. Scale bar: 5 μm.
Figure 3. PIP2 retains its colocalization with UBF during mitosis as demonstrated by near-native IEM. Immunogold electron microscopy of the cryo-immobilized and freeze-substituted mitotic cells demonstrates in ultrastructural detail that PIP2 colocalizes with UBF in discernible NORs (marked by arrowheads). PIP2 occupies the whole NOR volume rather than being sequestered in distinct sub-domains. Ch, chromatin; MT, microtubules. Scale bar: 200 nm.
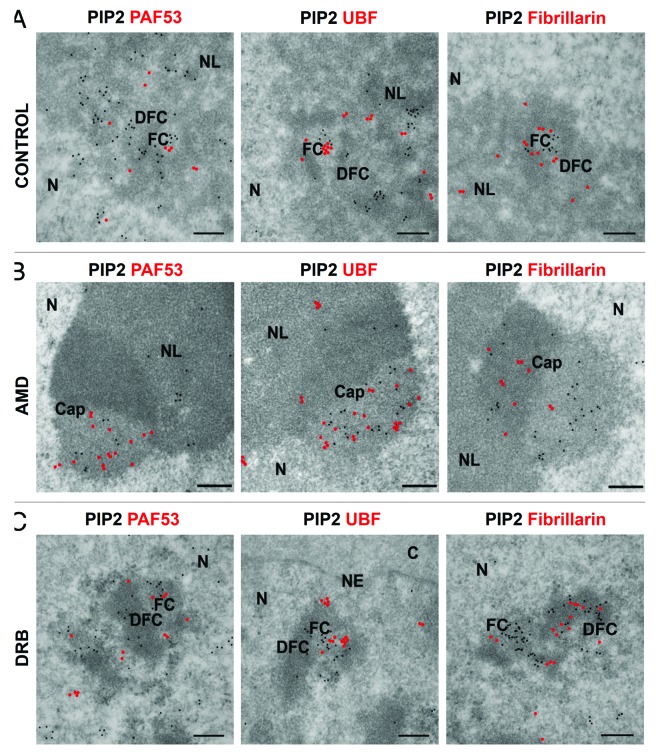
Low concentration of AMD was used to investigate the effect of chemically provoked inhibition of Pol I transcription on PIP2 colocalization with Pol I and UBF. In transcriptionally active interphase nucleoli, we showed by confocal microscopy that PIP2 colocalizes with Pol I and UBF, as seen by signal intensity profiles (Fig. 4A). In agreement with this, IEM also showed PIP2 localization in a close proximity to Pol I and UBF in the FC region of nucleoli, which is the counterpart of mitotic NORs, where the inactive components of Pol I transcription machinery accumulate during interphase (Fig. 5A). Pol I inhibition by AMD treatment results in nucleolar segregation and formation of a central body associated with caps.32 Immunofluorescence microscopy showed that PIP2 still colocalized with Pol I and UBF even though Pol I transcription was blocked (Fig. 4B). IEM further demonstrated intermingling clusters of PIP2 with Pol I and UBF in the light part of the caps as seen in Figure 5B.
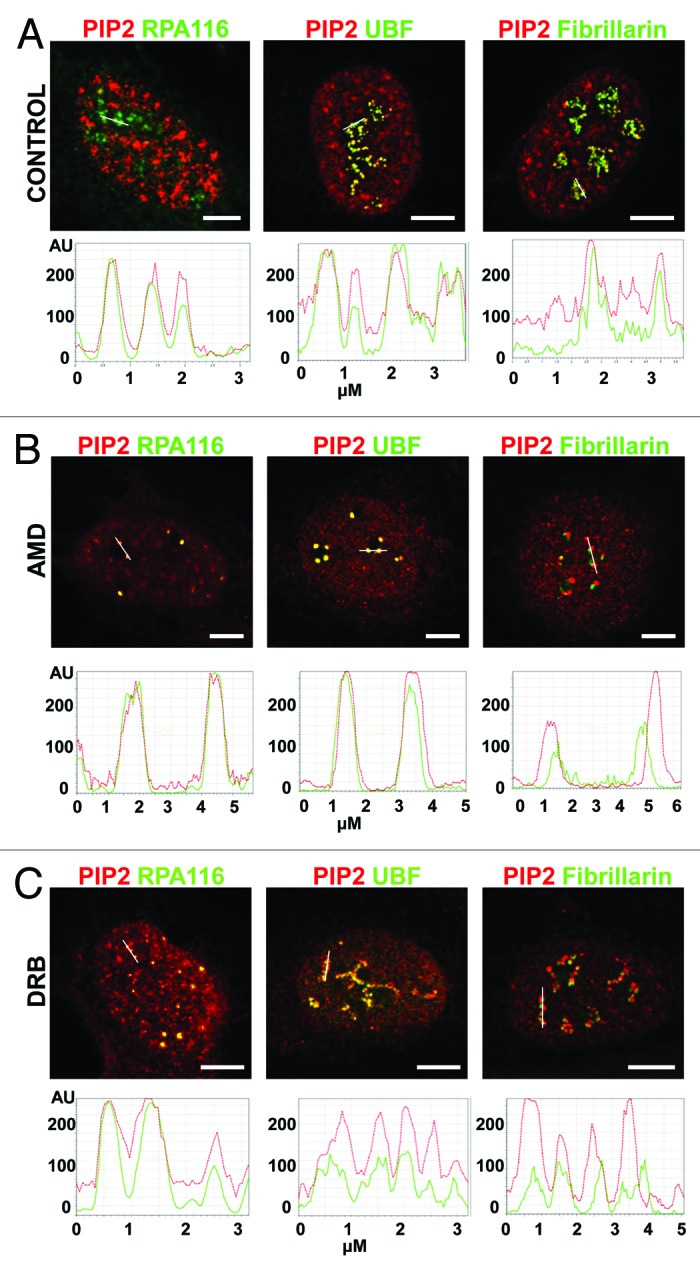
Figure 4. PIP2 colocalization with Pol I and UBF is not influenced by transcription inhibition while PIP2 colocalization with fibrillarin is disrupted during transcription inhibition as shown by confocal microscopy. (A) Immunocolocalization studies show the colocalization of PIP2 with Pol I, UBF, and fibrillarin in control cells as seen in the intensity profiles. (B) Pol I transcription inhibition by AMD did not affect the colocalization of PIP2 with Pol I and UBF. On the other hand, PIP2/fibrillarin colocalization was disrupted by Pol I transcription inhibition. (C) Pol II transcription inhibition by DRB did not change the colocalization of PIP2 with Pol I and UBF in nucleoli. On the other hand, PIP2-fibrillarin colocalization was lost upon inhibition of Pol II transcription. In graph: x-axis is in μm, y-axis is in arbitrary units. Scale bar: 5 μm.
Figure 5. PIP2 colocalization with Pol I and UBF is not influenced by transcription inhibition while PIP2 colocalization with fibrillarin is disrupted by transcription inhibition as shown by IEM. (A) IEM results show that PIP2 is in close proximity to Pol I in the nucleolus and colocalizes with UBF in the FC and with fibrillarin in DFC regions, respectively. (B) In AMD inhibited cells, PIP2 localizes to the light part of the caps together with Pol I and UBF while fibrillarin localizes mainly to the denser part of the caps after the inhibition of Pol I transcription. (C) Upon DRB treatment, PIP2 colocalizes with Pol I and UBF in the inner space of FCs as well as on the border between FC and DFC regions, while in the DFC, PIP2 and fibrillarin are arranged in a necklace-like manner. N, nucleus; NL, nucleolus; FC, fibrillar center; DFC, dense fibrillar component. Scale bar: 200 nm.
To prove that this effect is Pol I–specific and not caused by the side inhibition of Pol II transcription, we used DRB as an inhibitor of the phosphorylation of Pol II C-terminal domain (CTD), blocking its activity.33 DRB-induced Pol II inhibition causes alterations in the nucleolar architecture and results in “beads on a string” morphology. Despite the fact that there are prominent changes in the structure of nucleolar subdomains, rRNA genes remain transcriptionally active.34 Upon Pol II transcription inhibition, nucleolar beads were assembled and PIP2 colocalization with Pol I and UBF persisted as documented in the corresponding signal intensity profiles (Fig. 4C). In agreement, IEM demonstrated PIP2 localization in very close vicinity to Pol I and UBF in DRB-treated cells (Fig. 5C).
Taken together, these results show that the association of PIP2 with the components of Pol I pre-initiation complex is not dependent on active transcription, and it is maintained throughout the cell cycle.
PIP2 colocalization with fibrillarin in nucleoli is transcription-dependent
Fibrillarin methylates pre-rRNAs16 and its recruitment to the DFC region of the nucleolus is dependent on active transcription.35,36 We investigated the effect of transcription inhibition on the localization of PIP2 and fibrillarin in the DFC region of nucleoli. PIP2 and fibrillarin colocalize in actively transcribing cells (Fig. 4A), and PIP2/fibrillarin clusters can be readily seen in the DFC region by IEM (Fig. 5A). However, upon inhibition of Pol I transcription, PIP2/fibrillarin colocalization is lost as seen in the signal intensity profile (Fig. 4B). Fibrillarin is concentrated in the dense part of the caps, and in most cases does not colocalize with PIP2 after AMD treatment (Fig. 5B). Upon Pol II inhibition by DRB treatment, PIP2 colocalization with fibrillarin is restricted to a limited area in the nucleolus due to the segregation of DFC and FC regions in newly formed nucleolar beads (Fig. 4C). In accordance, we clearly distinguished DFC region, where PIP2 intermingled with fibrillarin, and FC region, where only PIP2 was localized (Fig. 5C).
These results indicate that PIP2-fibrillarin colocalization is dependent on active transcription.
Discussion
It has been known for many years that the nucleus retains significant amounts of lipids, including phospholipids,26 even after experimental removal of the nuclear membrane.37 Due to the lack of membranous structures inside the nucleus, it has been suggested that other hydrophobic molecules maintain a particular environment for phospholipids to be localized in the nucleus (for reviews see refs. 38 and 39). The nucleolus is not separated from the nucleoplasm by a membrane yet it has a unique protein-nucleic acid composition. Nucleolar proteins are rich in basic residues which enable them to translocate into the nucleolus upon binding to GTP protein.40 Therefore, due to favorable environment of basic proteins, the nucleolus can harbor acidic phospholipids such as PIP2. In spite of the fact that the structure and composition of the nucleolus has been intensively studied, the mechanisms of their formation and maintenance remain unclear (for review see ref. 41). Here we studied the dependence of subnucleolar localization of PIP2 and the proteins involved in pre-rRNA synthesis and processing on the transcriptional activity of the cells.
We found that PIP2 is not localized in the GC region, where the assembly and maturation of pre-ribosome particles take place, but it forms clusters in the FC and DFC regions responsible for transcription and processing of rRNA. Recently, we have demonstrated that PIP2 promotes Pol I transcription and directs UBF binding to a more selective site on the rDNA promoter. Furthermore, we showed the alteration in fibrillarin binding to rRNA upon interaction with PIP2.27 Since UBF and fibrillarin are reported to have roles in the formation of nucleoli,42,43 these results suggest a role for PIP2 as well in nucleoli formation. Here we investigated if PIP2 is still present in the sites of Pol I transcription factory following inhibition of transcription. The effect of transcription inhibition was tested in native conditions during mitosis as well as using the treatment with AMD and DRB, which alter nucleolar architecture significantly. Indeed, we found that upon either physiological or chemically induced inhibition of Pol I transcription, PIP2 maintains its association with the components of Pol I pre-initiation complex but not with fibrillarin. These data reinforce the view that PIP2 interacts with fibrillarin only upon active pre-rRNA transcription while its binding to Pol I complex is not dependent on the synthesis of rRNA. It is known that the transcription factors are shuttling between the NORs and the cytoplasm even during mitosis, and this trafficking depends on the mitotic stage.17 In spite of this, UBF owing to its DNA-binding capacity is indispensably associated with non-condensed fibers of rDNA during the cell cycle.17,20 According to the modified basic and hydrophobic (BH) scale developed by Brzeska et al.,44 UBF sequence includes potential lipid-binding sites. Three of them are located in the HMG box 1 and the forth is positioned in the HMG box 4. Interestingly, all four sites are distributed inside the DNA-binding sites. We suggest that PIP2 associated with UBF is engaged in the formation of the core part of rDNA helix structure thus maintaining the open chromatin state of NORs independent of the Pol I transcription. During mitosis, certain Pol I transcription factors are phosphorylated by the Cdk1-cyclin B kinase, which is a prerequisite for establishing and retaining the rDNA transcription in the repressed state.19,45-47 To switch on the rRNA synthetic activity at the exit from mitosis, UBF has to lose the mitosis-specific inhibitory phosphorylations and be activated by phosphorylation at Ser 484 by G1-specific Cdk4/cyclin D1 kinase and at Ser 388 by Cdk 2/cyclin E&A kinase.48,49 Our data suggest that the interaction between PIP2 and UBF is not governed by these general phosphorylation-dephosphorylation mechanisms, but it is preserved throughout the cell cycle, indicating a structural role for PIP2 in the formation and maintenance of nucleolar architecture. It has been shown that Pol I subunit PAF53 contacts UBF directly, and both PAF53 and RPA116 subunits remain associated with Pol I transcription machinery independent of the rRNA synthesis.50,51 By BH scale, two potential lipid-binding sites are positioned in PAF 53 sequence and one potential site is located in RPA 116 sequence. In accordance with these data, we showed that PAF53 and RPA116 retain the binding with UBF and PIP2 in the absence of Pol I transcription supporting the notion of PIP2 involvement in the core complex of NORs.
It is known that in yeast cells mutations impairing Pol I elongation also impair the cleavage of precursor rRNA, suggesting a connection between production and processing of rRNA.52,53 Our data that PIP2 interacts with fibrillarin only upon active pre-rRNA transcription, support the notion that coordination of Pol I transcription and pre-rRNA processing factors is mediated by the production of rRNA.
In summary, we showed the interaction of PIP2 with the indispensable components of Pol I pre-initiation complex regardless of Pol I transcription suggesting the role for PIP2 in the formation and maintenance of the core platform of rDNA helix structure. Thus, we identified for the first time a lipid component of the nucleo-protein NOR complex. Obviously, proving this idea will require further work on the alterations in NOR architecture, nucleolar structure, and ribosomal gene transcription upon depletion of nucleolar PIP2 in situ.
Material and Methods
Cell culture and inhibitors
Human osteosarcoma (U2OS) cells and cervical carcinoma (HeLa) cells were kept in DMEM with 10% fetal calf serum in 5% CO2/air, 37 °C, humidified atmosphere. Cells were treated with AMD (0.02 µg/ml) for 2 h or with DRB (50 µg/ml) for 1 h.
Plasmids and production of recombinant proteins
GST-tagged PLCδ1PH (1–140) was received from Dr Hitoshi Yagisawa.54 Recombinant PLCδ1PH domain is a commonly used PIP2 probe due to its high affinity to the head domain of PIP2 molecule.27,54 For GST-tagged PLCδ1PH, the purification was performed on glutathione-agarose column (G4510, Sigma Aldrich) which had been equilibrated with BC100 (20 mM Tris pH 8.0, 0.1 mM EDTA, 20% glycerol, 100 mM NaCl). After washes with BC100, 0.1% NP40, 1 mM DTT, and complete protease inhibitors cocktail (5056489001, Roche Diagnostics GmbH) proteins were eluted with 50 mM Tris-HCL, pH 8.0 having 0.1g of reduced L-Glutathione (G4251, Sigma Aldrich).
Cell fractionation and immunoprecipitation
For obtaining chromosomal fraction enriched with NORs, adherent HeLa cells were treated with nocodazole (20 ng/ml) for 6 h. Mitotic cells were harvested by shaking-off, lysed in the buffer (110 mM CH3COOK, 10 mM HEPES, pH 7.5, 2 mM MgCl2, 0.5% Brij98, complete protease inhibitor cocktail) and homogenized with a G22 needle. The resultant extract was centrifuged at 600 g for 5 min at 4 °C. The pellet was resuspended in the IP buffer (150 mM NaCl, 50 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 1% NP-40, complete protease inhibitor cocktail), sonicated and treated with Benzonase (CNBI70664, Millipore; 4000 units) for 1 h at 4 °C. After centrifugation at 16100 g for 20 min at 4 °C, supernatant was used in the immunoprecipitation assays. Samples were incubated with 2 µg of either anti-PIP2 mouse monoclonal IgM antibody (ZA045, Echelon Biosciences Inc) or mouse monoclonal IgM-isotype control (ab18401, Abcam) overnight at 4 °C, and then with 50 µl of protein L-agarose beads (20520, Thermo Scientific Pierce) for 2.5 h at 4 °C. Immunoprecipitates were thoroughly washed with the IP buffer, boiled in Laemmli sample buffer for 10 min, and separated by SDS-PAGE for western blot detection. NOR integrity was monitored with double immunolabeling of UBF and PIP2 or Pol I and PIP2 followed by confocal microscopy taking the samples during cell fractionation.
Immunoblot analysis
Proteins transferred to nitrocellulose membrane (Pall 66485, Pall Corporation) were blocked with 5% non-fat milk in PBS for 1 h and incubated with the appropriate specific antibodies diluted in 1% BSA in PBST overnight at 4 °C. Immunoblotting signals were detected with the appropriate IRDye donkey anti-rabbit or IRDye goat anti-rabbit antibodies and analyzed by Odyssey Infrared Imager 9120 (LI-COR Biosciences).
Light microscopy
Images were taken with either a confocal microscope (Leica TCS SP5 AOBS TANDEM) with 100 × (NA 1.4) oil immersion objective lens or a superresolution structured illumination microscope (ELYRA PS.1, Carl Zeiss; Andor iXon3 885 EMCCD camera, pixel size 8 × 8 µm) with Plan-Apochromat 63x/1.4 Oil DIC M27 oil immersion objective lens using the parameters as follows: number of SIM rotations = 5; SIM grating periods varied according to the excitation wavelength from 34.0 µm to 42.0 µm.
Immunoelectron microscopy
Interphase HeLa cells were fixed in 3% formaldehyde plus 0.1% glutaraldehyde and embedded into LR White resin by standard procedure.55 Mitotic HeLa cells were high-pressure frozen, freeze-substituted and embedded into LR White resin according to a previously published protocol.56 Thin sections (70 nm) were examined in a FEI Morgagni 268 transmission electron microscope at 80 kV and in a Tecnai G2 20 LaB6 electron microscope (FEI) at 200 kV. The images were captured with Mega View III CCD camera (pixel size 6.45 × 6.45 µm) and with Gatan Model 894 UltraScan 1000 camera (pixel size 14 × 14 µm). Multiple sections of at least three independent immunogold labeling experiments were analyzed. Adobe Photoshop CS3 Version 10.0 was used to identify the geometrical centers of 6 nm gold nanoparticles and then cover co-centrically with red dots to facilitate the visualization of these small nanoparticles in images.
Antibodies
Primary antibodies: anti-PIP2 mouse monoclonal IgM antibody (ab11039, Abcam; 4 µg/ml for SIM, 16 µg/ml for confocal microscopy, 32 µg/ml for IEM), anti-GST rabbit polyclonal antibody (gift from Dr Igor Shevelev; 5 µg/ml), anti-UBF rabbit polyclonal antibody (sc-9131, Santa Cruz Biotechnology; 0.4 µg/ml for immunoblotting), anti-UBF antibody from human autoimmune serum (gift from Dr Renate Voit; 1:1200 for SIM), anti-UBF rabbit polyclonal antibody (HPA006385, Sigma Aldrich; 2.6 µg/ml for IEM), anti-UBF mouse monoclonal IgG1 antibody (sc13125, Santa Cruz Biotechnology; 2 µg/ml for confocal microscopy), anti-PAF53 rabbit polyclonal antibody (gift from Prof Ingrid Grummt; 1:1000 for immunoblotting), anti-PAF53 mouse monoclonal IgG1 antibody (611413, BD Transduction Laboratories; 2.5 µg/ml for IEM), anti-RPA116 rabbit polyclonal antibody (gift from Prof Ingrid Grummt; 1:1000 for immunoblotting, 2 µg/ml for confocal microscopy), anti-fibrillarin rabbit monoclonal IgG antibody (2639, Cell Signaling Technology Inc.; 0.3 µg/ml for IEM), anti-fibrillarin mouse monoclonal IgG1 antibody (ab4566, Abcam; 1:100 for confocal microscopy). Secondary antibodies: IRDye 680RD donkey anti-rabbit IgG (H+L) (926-68073, LI-COR Biosciences; 1:10000), IRDye 800CW goat anti-rabbit IgG (H+L) (926-32211, LI-COR Biosciences; 1:10000), goat anti-mouse IgM conjugated with Alexa 555 (A21426, Invitrogen; 10 µg/ml for SIM), donkey anti-mouse IgM conjugated with Cy3 (715-165-140, Jackson ImmunoResearch; 10 µg/ml for confocal microscopy), donkey anti-mouse IgG conjugated with Alexa 488 (A21202, Invitrogen; 5 µg/ml for confocal microscopy), goat anti-rabbit IgG conjugated with Alexa 647 (A21245, Invitrogen; 5 µg/ml for confocal microscopy), goat anti-human IgG conjugated with Alexa 488 (A11013, Invitrogen; 5 µg/ml for SIM), goat anti-mouse IgM (µ-chain specific) antibody coupled with either 6 nm (115-195-075) or 12 nm (115-205-075) colloidal gold particles, goat anti-mouse IgG (H+L) antibody coupled with 6 nm colloidal gold particles (115-195-068), goat anti-rabbit IgG (H+L) antibody coupled with either 6 nm (111-195-144) or 12 nm (111-205-144) colloidal gold particles (Jackson ImmunoResearch Laboratories Inc); all gold-conjugated secondary antibodies were diluted 1:30.
Disclosure of Potential Conflicts of Interest
There are no conflicts of either financial or personal interest.
Acknowledgments
We thank Pavel Kříž, Iva Jelínková, and Ivana Nováková for their excellent technical assistance. We also thank to Prof Ingrid Grummt, Dr Igor Shevelev, and Dr Hitoshi Yagisawa for sharing plasmids and antibodies with us. We thank Prof William C Earnshaw, Ilona Kalasová, and Dr Tomáš Vacík for the methodological tips. We also thank Dr Jacques Paysan for his help with SIM. We thank Irina Studenyak for proofreading. This work was supported by the Grant Agency of the Czech Republic (P305/11/2232); the Ministry of Education, Youth and Sports of the Czech Republic (LD12063); the Ministry of Education, Youth and Sports, the European Regional Development Fund, and the Research and Development for Innovations Operational Programme (CZ.1.05/1.1.00/02.0109); CONACYT (176598); IMG institutional grant (RVO68378050).
Glossary
Abbreviations:
- NOR
nucleolar organizing region
- FC
fibrillar center
- DFC
dense fibrillar component
- GC
granular component
- rDNA
ribosomal DNA
- Pol I
RNA polymerase I
- UBF
upstream binding factor
- HMG
high mobility group
- PIC
pre-initiation complex
- SL1
promoter selectivity factor
- TBP
TATA-binding protein
- TAF
TBP-associated factors
- rRNA
ribosomal RNA
- snoRNA
small nucleolar RNA
- PIP2
phosphatidylinositol 4,5-bisphosphate
- PLC
phospholipase C
- IP3
inositol 1,4,5-trisphosphate
- DAG
diacylglycerol
- AMD
actinomycin D
- DRB
5,6-dichloro-1β-d-ribofuranosyl-benzimidazole
- SIM
super-resolution structured illumination microscopy
- HPF
high-pressure freezing
- FS
freeze-substitution
- IEM
immunoelectron microscopy
- CTD
Pol II C-terminal domain
- BH scale
basic and hydrophobic scale
- U2OS cells
human osteosarcoma cells
- HeLa cells
cervical carcinoma cells
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/27154
References
- 1.Boisvert FM, van Koningsbruggen S, Navascués J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–85. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 2.Pederson T, Tsai RY. In search of nonribosomal nucleolar protein function and regulation. J Cell Biol. 2009;184:771–6. doi: 10.1083/jcb.200812014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bazett-Jones DP, Leblanc B, Herfort M, Moss T. Short-range DNA looping by the Xenopus HMG-box transcription factor, xUBF. Science. 1994;264:1134–7. doi: 10.1126/science.8178172. [DOI] [PubMed] [Google Scholar]
- 4.Stefanovsky VY, Pelletier G, Bazett-Jones DP, Crane-Robinson C, Moss T. DNA looping in the RNA polymerase I enhancesome is the result of non-cooperative in-phase bending by two UBF molecules. Nucleic Acids Res. 2001;29:3241–7. doi: 10.1093/nar/29.15.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stefanovsky V, Langlois F, Gagnon-Kugler T, Rothblum LI, Moss T. Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodeling. Mol Cell. 2006;21:629–39. doi: 10.1016/j.molcel.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 6.Kermekchiev M, Workman JL, Pikaard CS. Nucleosome binding by the polymerase I transactivator upstream binding factor displaces linker histone H1. Mol Cell Biol. 1997;17:5833–42. doi: 10.1128/mcb.17.10.5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell SP, Learned RM, Jantzen HM, Tjian R. Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science. 1988;241:1192–7. doi: 10.1126/science.3413483. [DOI] [PubMed] [Google Scholar]
- 8.Comai L, Tanese N, Tjian R. The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell. 1992;68:965–76. doi: 10.1016/0092-8674(92)90039-F. [DOI] [PubMed] [Google Scholar]
- 9.Kwon H, Green MR. The RNA polymerase I transcription factor, upstream binding factor, interacts directly with the TATA box-binding protein. J Biol Chem. 1994;269:30140–6. [PubMed] [Google Scholar]
- 10.Beckmann H, Chen JL, O’Brien T, Tjian R. Coactivator and promoter-selective properties of RNA polymerase I TAFs. Science. 1995;270:1506–9. doi: 10.1126/science.270.5241.1506. [DOI] [PubMed] [Google Scholar]
- 11.Heix J, Grummt I. Species specificity of transcription by RNA polymerase I. Curr Opin Genet Dev. 1995;5:652–6. doi: 10.1016/0959-437X(95)80035-2. [DOI] [PubMed] [Google Scholar]
- 12.O’Sullivan AC, Sullivan GJ, McStay B. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol Cell Biol. 2002;22:657–68. doi: 10.1128/MCB.22.2.657-668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Copenhaver GP, Putnam CD, Denton ML, Pikaard CS. The RNA polymerase I transcription factor UBF is a sequence-tolerant HMG-box protein that can recognize structured nucleic acids. Nucleic Acids Res. 1994;22:2651–7. doi: 10.1093/nar/22.13.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denissov S, Lessard F, Mayer C, Stefanovsky V, van Driel M, Grummt I, Moss T, Stunnenberg HG. A model for the topology of active ribosomal RNA genes. EMBO Rep. 2011;12:231–7. doi: 10.1038/embor.2011.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayer C, Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25:6384–91. doi: 10.1038/sj.onc.1209883. [DOI] [PubMed] [Google Scholar]
- 16.Hernandez-Verdun D. The nucleolus today. J Cell Sci. 1991;99:465–71. doi: 10.1242/jcs.99.3.465. [DOI] [PubMed] [Google Scholar]
- 17.Chen D, Dundr M, Wang C, Leung A, Lamond A, Misteli T, Huang S. Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins. J Cell Biol. 2005;168:41–54. doi: 10.1083/jcb.200407182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roussel P, André C, Comai L, Hernandez-Verdun D. The rDNA transcription machinery is assembled during mitosis in active NORs and absent in inactive NORs. J Cell Biol. 1996;133:235–46. doi: 10.1083/jcb.133.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sirri V, Roussel P, Hernandez-Verdun D. The mitotically phosphorylated form of the transcription termination factor TTF-1 is associated with the repressed rDNA transcription machinery. J Cell Sci. 1999;112:3259–68. doi: 10.1242/jcs.112.19.3259. [DOI] [PubMed] [Google Scholar]
- 20.Leung AK, Gerlich D, Miller G, Lyon C, Lam YW, Lleres D, Daigle N, Zomerdijk J, Ellenberg J, Lamond AI. Quantitative kinetic analysis of nucleolar breakdown and reassembly during mitosis in live human cells. J Cell Biol. 2004;166:787–800. doi: 10.1083/jcb.200405013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sirri V, Hernandez-Verdun D, Roussel P. Cyclin-dependent kinases govern formation and maintenance of the nucleolus. J Cell Biol. 2002;156:969–81. doi: 10.1083/jcb.200201024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martelli AM, Manzoli L, Faenza I, Bortul R, Billi A, Cocco L. Nuclear inositol lipid signaling and its potential involvement in malignant transformation. Biochim Biophys Acta. 2002;1603:11–7. doi: 10.1016/s0304-419x(02)00066-5. [DOI] [PubMed] [Google Scholar]
- 23.Mellman DL, Gonzales ML, Song C, Barlow CA, Wang P, Kendziorski C, Anderson RAA. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature. 2008;451:1013–7. doi: 10.1038/nature06666. [DOI] [PubMed] [Google Scholar]
- 24.Okada M, Jang SW, Ye K. Akt phosphorylation and nuclear phosphoinositide association mediate mRNA export and cell proliferation activities by ALY. Proc Natl Acad Sci U S A. 2008;105:8649–54. doi: 10.1073/pnas.0802533105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osborne SL, Thomas CL, Gschmeissner S, Schiavo G. Nuclear PtdIns(4,5)P2 assembles in a mitotically regulated particle involved in pre-mRNA splicing. J Cell Sci. 2001;114:2501–11. doi: 10.1242/jcs.114.13.2501. [DOI] [PubMed] [Google Scholar]
- 26.Fraschini A, Biggiogera M, Bottone MG, Martin TE. Nuclear phospholipids in human lymphocytes activated by phytohemagglutinin. Eur J Cell Biol. 1999;78:416–23. doi: 10.1016/S0171-9335(99)80084-3. [DOI] [PubMed] [Google Scholar]
- 27.Yildirim S, Castano E, Sobol M, Philimonenko VV, Dzijak R, Venit T, Hozák P. Involvement of phosphatidylinositol 4,5-bisphosphate in RNA polymerase I transcription. J Cell Sci. 2013;126:2730–9. doi: 10.1242/jcs.123661. [DOI] [PubMed] [Google Scholar]
- 28.Sirri V, Urcuqui-Inchima S, Roussel P, Hernandez-Verdun D. Nucleolus: the fascinating nuclear body. Histochem Cell Biol. 2008;129:13–31. doi: 10.1007/s00418-007-0359-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panse SL, Masson C, Héliot L, Chassery JM, Junéra HR, Hernandez-Verdun D. 3-D organization of ribosomal transcription units after DRB inhibition of RNA polymerase II transcription. J Cell Sci. 1999;112:2145–54. doi: 10.1242/jcs.112.13.2145. [DOI] [PubMed] [Google Scholar]
- 30.Sobol MA, Philimonenko VV, Philimonenko AA, Hozák P. Quantitative evaluation of freeze-substitution effects on preservation of nuclear antigens during preparation of biological samples for immunoelectron microscopy. Histochem Cell Biol. 2012;138:167–77. doi: 10.1007/s00418-012-0931-6. [DOI] [PubMed] [Google Scholar]
- 31.Gébrane-Younès J, Fomproix N, Hernandez-Verdun D. When rDNA transcription is arrested during mitosis, UBF is still associated with non-condensed rDNA. J Cell Sci. 1997;110:2429–40. doi: 10.1242/jcs.110.19.2429. [DOI] [PubMed] [Google Scholar]
- 32.Puvion-Dutilleul F, Puvion E, Bachellerie JP. Early stages of pre-rRNA formation within the nucleolar ultrastructure of mouse cells studied by in situ hybridization with a 5’ETS leader probe. Chromosoma. 1997;105:496–505. doi: 10.1007/BF02510486. [DOI] [PubMed] [Google Scholar]
- 33.Bird G, Zorio DA, Bentley DL. RNA polymerase II carboxy-terminal domain phosphorylation is required for cotranscriptional pre-mRNA splicing and 3′-end formation. Mol Cell Biol. 2004;24:8963–9. doi: 10.1128/MCB.24.20.8963-8969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scheer U, Hügle B, Hazan R, Rose KM. Drug-induced dispersal of transcribed rRNA genes and transcriptional products: immunolocalization and silver staining of different nucleolar components in rat cells treated with 5,6-dichloro-beta-D-ribofuranosylbenzimidazole. J Cell Biol. 1984;99:672–9. doi: 10.1083/jcb.99.2.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dundr M, Meier UT, Lewis N, Rekosh D, Hammarskjöld ML, Olson MO. A class of nonribosomal nucleolar components is located in chromosome periphery and in nucleolus-derived foci during anaphase and telophase. Chromosoma. 1997;105:407–17. doi: 10.1007/BF02510477. [DOI] [PubMed] [Google Scholar]
- 36.Kopp K, Gasiorowski JZ, Chen D, Gilmore R, Norton JT, Wang C, Leary DJ, Chan EK, Dean DA, Huang S. Pol I transcription and pre-rRNA processing are coordinated in a transcription-dependent manner in mammalian cells. Mol Biol Cell. 2007;18:394–403. doi: 10.1091/mbc.E06-03-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vann LR, Wooding FB, Irvine RF, Divecha N. Metabolism and possible compartmentalization of inositol lipids in isolated rat-liver nuclei. Biochem J. 1997;327:569–76. doi: 10.1042/bj3270569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Irvine RF. Nuclear lipid signalling. Nat Rev Mol Cell Biol. 2003;4:349–60. doi: 10.1038/nrm1100. [DOI] [PubMed] [Google Scholar]
- 39.Irvine RF. Nuclear inositide signalling -- expansion, structures and clarification. Biochim Biophys Acta. 2006;1761:505–8. doi: 10.1016/j.bbalip.2006.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai RY, McKay RD. A multistep, GTP-driven mechanism controlling the dynamic cycling of nucleostemin. J Cell Biol. 2005;168:179–84. doi: 10.1083/jcb.200409053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hernandez-Verdun D. Assembly and disassembly of the nucleolus during the cell cycle. Nucleus. 2011;2:189–94. doi: 10.4161/nucl.2.3.16246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fomproix N, Gébrane-Younès J, Hernandez-Verdun D. Effects of anti-fibrillarin antibodies on building of functional nucleoli at the end of mitosis. J Cell Sci. 1998;111:359–72. doi: 10.1242/jcs.111.3.359. [DOI] [PubMed] [Google Scholar]
- 43.Mais C, Wright JE, Prieto JL, Raggett SL, McStay B. UBF-binding site arrays form pseudo-NORs and sequester the RNA polymerase I transcription machinery. Genes Dev. 2005;19:50–64. doi: 10.1101/gad.310705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brzeska H, Guag J, Remmert K, Chacko S, Korn ED. An experimentally based computer search identifies unstructured membrane-binding sites in proteins: application to class I myosins, PAKS, and CARMIL. J Biol Chem. 2010;285:5738–47. doi: 10.1074/jbc.M109.066910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gavet O, Pines J. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev Cell. 2010;18:533–43. doi: 10.1016/j.devcel.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heix J, Vente A, Voit R, Budde A, Michaelidis TM, Grummt I. Mitotic silencing of human rRNA synthesis: inactivation of the promoter selectivity factor SL1 by cdc2/cyclin B-mediated phosphorylation. EMBO J. 1998;17:7373–81. doi: 10.1093/emboj/17.24.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sirri V, Roussel P, Hernandez-Verdun D. In vivo release of mitotic silencing of ribosomal gene transcription does not give rise to precursor ribosomal RNA processing. J Cell Biol. 2000;148:259–70. doi: 10.1083/jcb.148.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Voit R, Grummt I. Phosphorylation of UBF at serine 388 is required for interaction with RNA polymerase I and activation of rDNA transcription. Proc Natl Acad Sci U S A. 2001;98:13631–6. doi: 10.1073/pnas.231071698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Voit R, Hoffmann M, Grummt I. Phosphorylation by G1-specific cdk-cyclin complexes activates the nucleolar transcription factor UBF. EMBO J. 1999;18:1891–9. doi: 10.1093/emboj/18.7.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanada K, Song CZ, Yamamoto K, Yano K, Maeda Y, Yamaguchi K, Muramatsu M. RNA polymerase I associated factor 53 binds to the nucleolar transcription factor UBF and functions in specific rDNA transcription. EMBO J. 1996;15:2217–26. [PMC free article] [PubMed] [Google Scholar]
- 51.Seither P, Zatsepina O, Hoffmann M, Grummt I. Constitutive and strong association of PAF53 with RNA polymerase I. Chromosoma. 1997;106:216–25. doi: 10.1007/s004120050242. [DOI] [PubMed] [Google Scholar]
- 52.Schneider DA, French SL, Osheim YN, Bailey AO, Vu L, Dodd J, Yates JR, Beyer AL, Nomura M. RNA polymerase II elongation factors Spt4p and Spt5p play roles in transcription elongation by RNA polymerase I and rRNA processing. Proc Natl Acad Sci U S A. 2006;103:12707–12. doi: 10.1073/pnas.0605686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schneider DA, Michel A, Sikes ML, Vu L, Dodd JA, Salgia S, Osheim YN, Beyer AL, Nomura M. Transcription elongation by RNA polymerase I is linked to efficient rRNA processing and ribosome assembly. Mol Cell. 2007;26:217–29. doi: 10.1016/j.molcel.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yagisawa H, Sakuma K, Paterson HF, Cheung R, Allen V, Hirata H, Watanabe Y, Hirata M, Williams RL, Katan M. Replacements of single basic amino acids in the pleckstrin homology domain of phospholipase C-delta1 alter the ligand binding, phospholipase activity, and interaction with the plasma membrane. J Biol Chem. 1998;273:417–24. doi: 10.1074/jbc.273.1.417. [DOI] [PubMed] [Google Scholar]
- 55.Sobol M, Philimonenko VV, Hozák P. Comparison of methods of high-pressure freezing and automated freeze-substitution of suspension cells combined with LR White embedding. Histochem Cell Biol. 2010;134:631–41. doi: 10.1007/s00418-010-0757-z. [DOI] [PubMed] [Google Scholar]
- 56.Sobol M, Nebesářová J, Hozák P. A method for preserving ultrastructural properties of mitotic cells for subsequent immunogold labeling using low-temperature embedding in LR White resin. Histochem Cell Biol. 2011;135:103–10. doi: 10.1007/s00418-010-0771-1. [DOI] [PubMed] [Google Scholar]