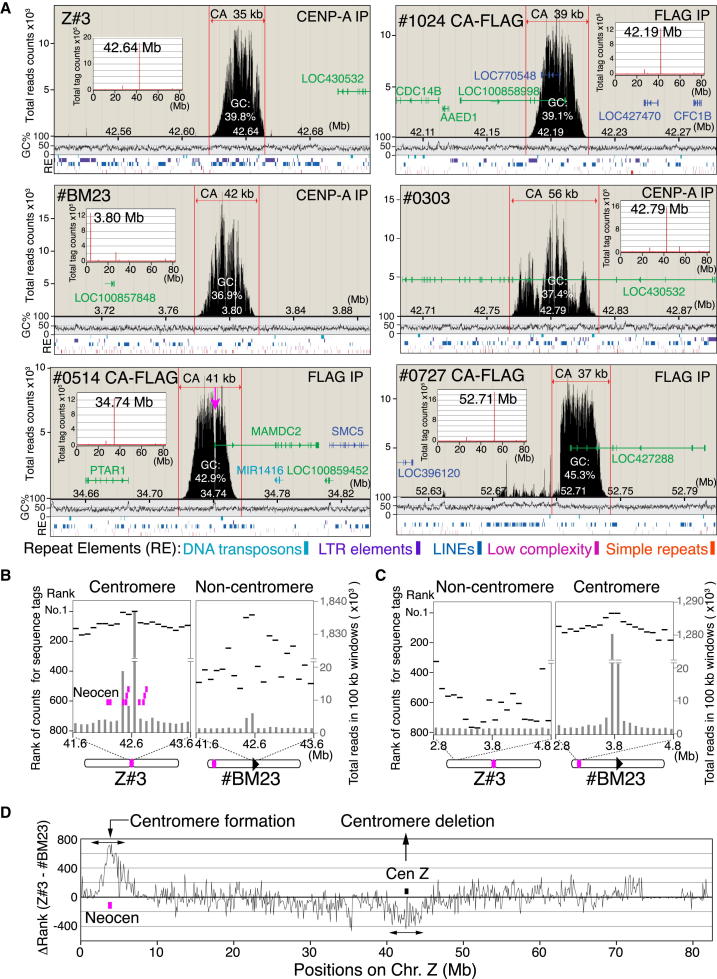
Figure 3.
Neocentromeres Are ∼40 kb Long
(A) ChIP-seq analysis with anti-FLAG or anti-CENP-A antibodies in cells containing neocentromere. Size of the CENP-A domain in each cell line was shown. We used anti-FLAG antibodies for cells expressing CENP-A-FLAG (FLAG-IP). For cells not expressing CENP-A-FLAG, we used native CENP-A antibodies (CENP-A-IP). IP DNAs were deeply sequenced and sequence data were mapped on chicken genome database. We first identified a major peak as a neocentromere (each position is indicated) from entire chromosome Z in each cell line and examined detail distribution around the peak with a higher resolution. GC% contents and distribution of transposons, repeat sequence, and genes in CENP-A-associated DNAs are shown. Arrow in data of #0514 cells indicates a gap of CENP-A distribution, which corresponds to exon 1 of MAMDC2 gene (also see Figure 7).
(B) Counts of sequence reads (gray bar) for CENP-A-IP DNAs around centromere (Z#3) and noncentromere (#BM23) region. Ranking for these counts are also shown (black line). Pink dots are shown as neocentromere loci. CENP-A-associated DNAs were enriched in 2 Mb around the major CENP-A peak (Z#3). However, numbers of sequence tags associated with CENP-A were reduced in this region, when an endogenous centromere was removed (#BM23).
(C) Counts of sequence reads (gray bar) for CENP-A-IP and their ranking (black line) around centromere (#BM23) and noncentromere (Z#3) region. CENP-A cluster was observed around centromere (#BM23).
(D) Genome-wide ranking of numbers of sequence tags associated with CENP-A for Z#3 cells were subtracted from that for #BM23. A CENP-A cluster was observed around neocentromere region of #BM23.
See also Figures S3 and S4.