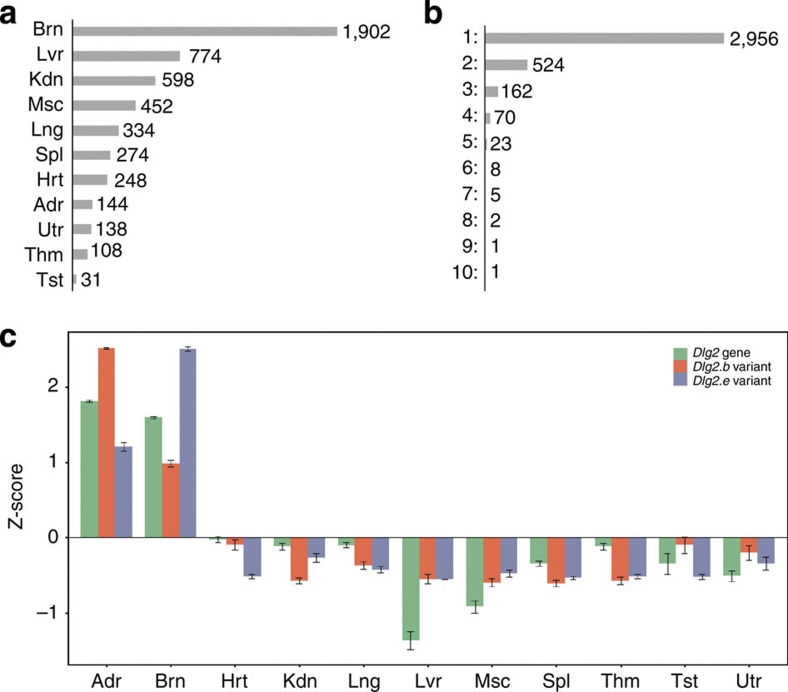
Figure 5. Organ-enriched alternatively spliced transcript expression.
(a) Numbers of organ-specific transcript variants for both female and male rats across the four developmental stages. A transcript was considered an organ-specific alternatively spliced variant if it had an FC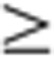 2 plus a Bonferroni-corrected P-value
2 plus a Bonferroni-corrected P-value 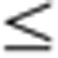 0.05 when compared with its expression in other organs across the four developmental stages. (b) Organ-specific transcript variant abundance. Distribution of genes (on the right) with a given number of variants (1 thru 10). (c) Organ-specific transcript variant expression for Dlg2. The y axis represents normalized (Mean=0 and s.d.=1) expression profile (mean±s.e., N=32 (or 16 for Tst and Utr)) Z-score and the x axis represents Dlg2 gene (green), Dlg2.b variant (orange) and Dlg2.e variant (blue) expression in each organ. Organs tested are: Adr, adrenal; Brn, brain; Hrt, heart; Kdn, kidney; Lng, lung; Lvr, liver; Msc, skeletal muscle; Spl, spleen; Thm, thymus; Tst, testis; and Utr, uterus.
0.05 when compared with its expression in other organs across the four developmental stages. (b) Organ-specific transcript variant abundance. Distribution of genes (on the right) with a given number of variants (1 thru 10). (c) Organ-specific transcript variant expression for Dlg2. The y axis represents normalized (Mean=0 and s.d.=1) expression profile (mean±s.e., N=32 (or 16 for Tst and Utr)) Z-score and the x axis represents Dlg2 gene (green), Dlg2.b variant (orange) and Dlg2.e variant (blue) expression in each organ. Organs tested are: Adr, adrenal; Brn, brain; Hrt, heart; Kdn, kidney; Lng, lung; Lvr, liver; Msc, skeletal muscle; Spl, spleen; Thm, thymus; Tst, testis; and Utr, uterus.