Fig. 1.
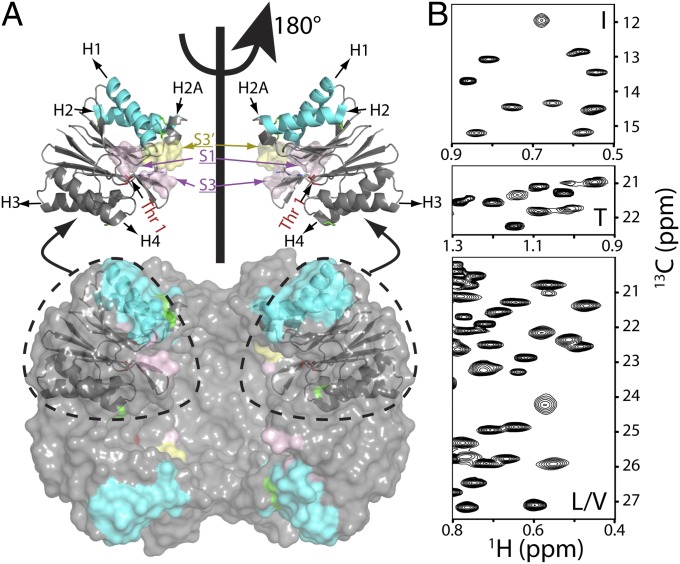
Near-complete I, L, M, T, and V methyl chemical shift assignments for HslV. (A) Surface model showing the dodecameric structure of HslV, comprising a pair of hexameric rings that are aligned coaxially (9) (Protein Data Bank ID code 1G3K) with the front four subunits removed to show the proteolytic chamber. All monomers of the hexamer are equivalent. Shown in Upper are ribbon diagrams for a pair of monomers, corresponding to the two front-facing subunits of the surface model, related to each other by a 180° rotation about an axis of sixfold symmetry. Helices are labeled H1, H2 (highlighted in cyan), H2A, H3, and H4, with catalytic residue T1 in red. Parts of the substrate binding pocket (S1, S3, and S3′ from an adjacent molecule) are also indicated. (B) Regions of 13C,1H HMQC spectra of [U-2H; Ileδ1-13CH3; Leu,Val-13CH3/12CD3; Met-13CH3]– or [U-2H; Ileδ1-13CH3; Met-13CH3; Thr-13CH3]–labeled HslV (18.8T and 40 °C).