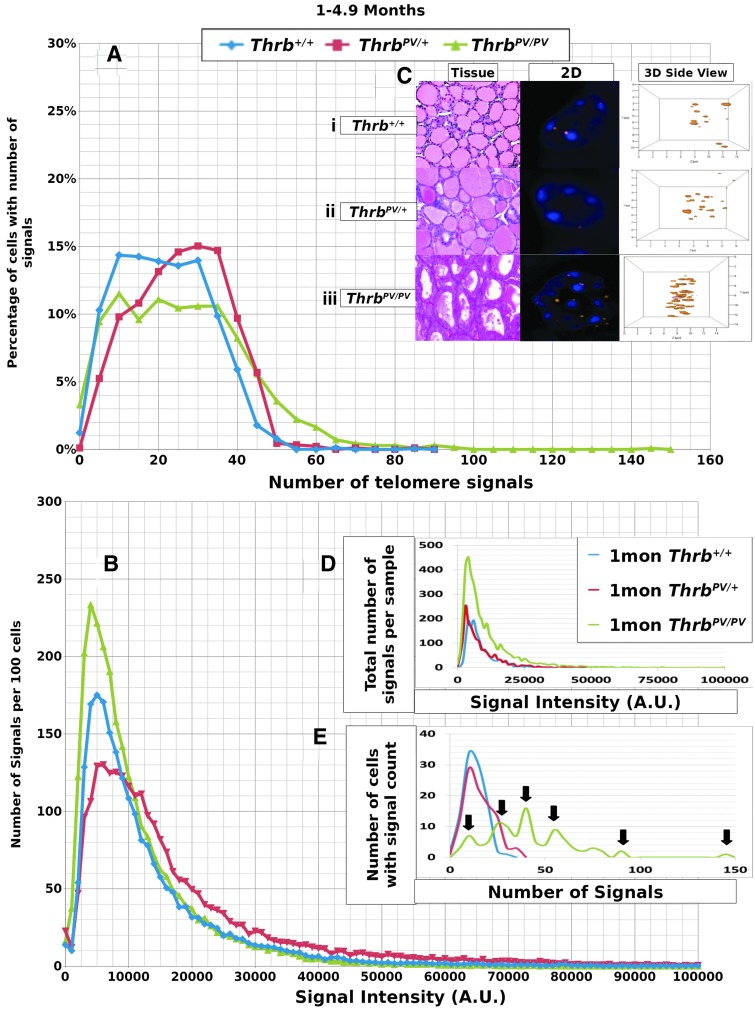
FIG. 2.
Data for 1–4.9 month old mouse thyrocytes. (A) y-Axis indicates the percentage of cells in the given genotypes that have the number of signals shown on the x-axis. This percentage was determined by dividing the number of cells with a specific number of signals by the total number of cells observed for each particular genotype. A shift to the right was observed in the ThrbPV/+ and especially the ThrbPV/PV genotypes and indicates cells with higher numbers of signals. (B) y-Axis indicates the number of signals per 100 cells observed at an intensity shown on the x-axis in arbitrary units (AU). This number was determined by dividing the number of signals observed by the total number of thyroid samples observed for each genotype. A shift upward indicates more signals of lesser intensity for ThrbPV/+ and ThrbPV/PV genotypes, and a shift to the right indicates more signals of higher intensity for ThrbPV/PV. (C) Representative thyroid areas (H&E stained) and individual thyrocyte nuclei selected from each genotype, DAPI nuclear stain with rhodamine-labelled telomere signals, (i) homozygous normal Thrb+/+, (ii) heterozygous ThrbPV/+ mutant, (iii) homozygous ThrbPV/PV mutant, shown as both 2D and 3D side nuclear images of telomere signals. Nuclei with more telomere signals are seen in nuclei of ThrbPV/PV thyrocytes. (D) Data from representative thyroid samples from mice of each genotype displaying signal intensity data. (E) Data from representative thyroid samples of mice for each genotype showing the number of signals per thyrocyte. Arrows indicate separate populations of cells with differing numbers of telomeres in the multipeaked ThrbPV/PV example.