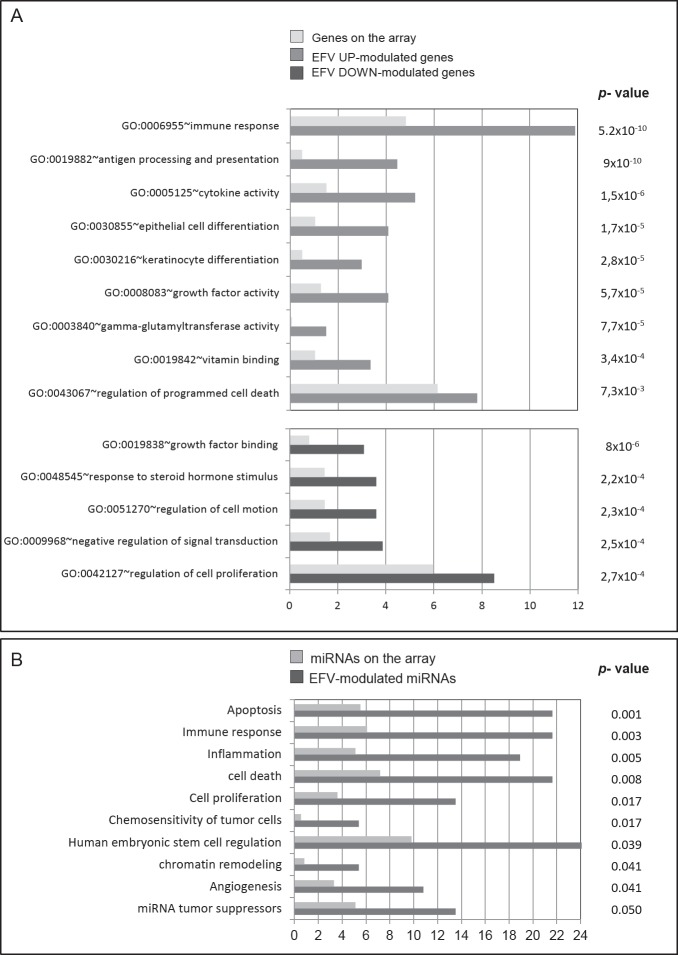
Figure 3. Gene ontology classification of EFV-modulated genes and miRNAs.
A) Gene Ontology classification of up- (gray histograms) and downregulated (dark histograms) genes in EFV-exposed A-375 cell cultures (at least 2.0-fold change, unpaired T-test P-value<0.05), using the David software. Biological process and Molecular function classes in which the genes fall are ranked according to the percentage of genes fitting each class relative to their expected frequency by chance (light gray histograms), calculated on the basis of the global array composition. The P value from the modified Fisher test classes enrichment (p < 0.05) is shown. B) Gene Ontology classification of differentially expressed miRNAs in EFV-exposed A-375 cell cultures (2.0-fold change, unpaired T-test P-value<0.05), using the Tool for annotations of human miRNAs (TAM, http://cmbi.bjmu.edu.cn/tam). The fold-enrichment values shows the overrepresentation of biological functions involving EFV-sensitive miRNAs (including up- and downregulated miRNAs), expressed as the pecentage of miRNAs fitting each class (gray histograms) relative to their expected frequency by chance, calculated on the basis of the global composition of the array (light gray histograms).