Abstract
Introduction
Vorinostat is a small molecule inhibitor of class I and II histone deacetylase (HDAC) enzymes which alters expression of target genes including the cell cycle gene p21, leading to cell cycle arrest and apoptosis.
Methods
Patients enrolled in a phase I trial were treated with vorinostat alone on day 1 and vorinostat and bortezomib in combination on day 9. Paired biopsies were obtained in eleven subjects. Blood samples were obtained on days 1 and 9 of cycle 1 prior to dosing, and 2 hours and 6 hours post dosing in all 60 subjects. Gene expression of p21, HSP70, AKT, nur77, ERB1 and ERB2 were evaluated in peripheral blood mononuclear cells and tissue samples. Chromatin immunoprecipitation of p21, HSP70 and Nur77 was also performed in biopsy samples.
Results
In peripheral blood mononuclear cells, Nur77 was significantly and consistently decreased two hours after vorinostat administration on both days 1 and 9, median ratio of gene expression relative to baseline of 0.69 with interquartile range (IQR) 0.49-1.04 (p<0.001); 0.28 (0.15-0.7) (p<0.001), respectively, with more pronounced decrease on day 9, when patients received both vorinostat and bortezomib. p21, a downstream target of nur77, was significantly decreased on day 9 two hours and six hours after administration of vorinostat and bortezomib, 0.67 (0.41-1.03) (p<0.01); 0.44 (0.25-1.3) (p<0.01), respectively. The ChiP assay demonstrateed a protein DNA interaction, in this case interaction of Nur77, HSP70 and p21 with acetylated histone H3, at baseline and at day 9 after treatment with vorinostat in tissue biopsies in most patients.
Conclusion
Vorinostat inhibits Nur77 expression, which in turn may decrease p21 and AKT expression in PBMCs. The influence of vorinostat on target gene expression in tumor tissue was variable, however, most patients demonstrated interaction of acetylated H3 with Nur77, HSP70 and p21 which provides evidence of interaction with the transcriptions active acetylated H3.
Keywords: SAHA, vorinostat, PS-341, bortezomib, phase I
INTRODUCTION
Histone deacetylation plays a critical role in the epigenetic regulation of gene expression as chromatin structure can influence gene expression (1). Condensed DNA that is wound around non-acetylated histones is transcriptionally inactive, while acetylated histone induces a conformational change that allows transcription factors access to DNA and allows transcriptional activity to occur. An interplay between histone acetyl-transferases (HATs) and histone deacetylases (HDACs) participates in the interconversion between permissive and repressive chromatin structures, modulating transcription regulating factor binding to the DNA template. Histone acetyl-transferases promote transcriptional activity by catalyzing the acetylation of N-terminal histone lysine residues while HDAC activity results in chromatin condensation and silencing of various genes. HDAC-dependent silencing of gene repressor can also result in increased transcription of other genes (2).
Vorinostat (suberoylanilide hydroxamic acid (SAHA) or MK-0683, Zolinza®, Merck, Whitehouse Station, NJ) is a small molecule inhibitor of class I and II HDAC enzymes, which enhances gene express of target genes. Vorinostat is currently approved for the treatment of cutaneous T cell lymphoma and has been shown in vitro to promote cell cycle arrest and apoptosis (3). Vorinostat causes upregulation of the cell cycle gene p21, leading to cell cycle arrest and inhibiting differentiation (4,5) in cell lines. Conversely, AKT, a downstream target of p21, has been shown to be down regulated by vorinostat (6).
Nur77 is a transcriptional factor in the nuclear receptor superfamily, which is induced by growth factors and mitogens, and its overexpression is associated with growth inhibition and apoptosis. Although Nur77 is induced by a variety of factors, very few target genes for Nur77 have been identified. However, Yoon and colleagues recently demonstrated that p21 induction was Nur77 dependent in a pancreas cancer cell model (7).
In addition, a number of nonhistone substrates have been identified and implicated in the antitumor activities of HDAC inhibitors, including molecular chaperones, such as heat shock protein 70 (HSP70), which is induced after treatment with HDAC inhibitors (8)), and avian erythroblastosis oncogene B (ErbB), which is down regulated by vorinostat (9).
In vitro studies have also demonstrated synergistic activity between vorinostat and bortezomib (10,11). Bortezomib may interact with vorinostat by disrupting multiple cyto-protective signaling pathways and by shifting the balance toward their stress-related counterparts, which may trigger mitochondrial damage, caspase activation, p21 and eventually lead to irreversible engagement of the apoptotic cascade.
The purpose of this study was to evaluate the in vivo influence of vorinostat alone and in combination with bortezomib on known apoptotic targets.
METHODS
Dosing
In this study, vorinostat was administered on two dose escalation schedules, Step A and Step B, respectively. In Step A, vorinostat (400 mg daily) was administered orally on days 1-14. During cycle 1, increasing doses of bortezomib (0.7-1.5mg/m2) were administered as an IV bolus on days 2, 5, 9 and 12. This dosing schedule was to allow evaluation of gene expression induction by vorinostat alone (Day 1) and in combination with bortezomib (Day 9). In Step B, vorinostat (200-300 mg) was administered orally twice daily on days 1-4 and 8-11 along with increasing doses of bortezomib (1 −1.3mg/m2).
Sample Collection
Paired biopsies were obtained in 11 subjects enrolled in the phase I trial combining vorinostat and bortezomib conducted at the University of Wisconsin. Core needle biopsies were obtained at baseline and within 2 hours of vorinostat dosing on day 1 and day 9 of cycle 1. Biopsies were flash frozen in liquid nitrogen and stored at −70 °C until analysis. Blood samples were also obtained in RNA Paxgene tubes on days 1 and 9 of cycle 1 prior to dosing, and 2 hours and 6 hours post dosing for the analysis of gene expression in all 60 subjects. Samples were stored at −70 °C until analysis.
The Health Sciences Institutional Review Board of the University of Wisconsin approved this trial prior to their implementation, and all patients gave informed written consent.
Chromatin Immunoprecipitation
Chromatin Immunoprecipitation (ChIP) using the anti-human acetyl-histone H3 (K9/K14) antibody was performed on tumor tissue biopsies with the ExactaCHIPTM Human acetyl-Histone H3(K9/K14) Chromatin Immunoprecipitation Kit from R&D systems as recommended by the manufacturer (12). This ExactaChIP kit is designed to immunoprecipitate acetyl-histone H3 (K9/K14)-DNA complexes and to allow for the rapid PCR detection of acetyl-histone H3 (K9/K14)-bound DNA fragments. Protein-DNA complexes were fixed by formaldehyde crosslinking, the chromatin was sheared by sonication, and the protein of interest was immunoprecipitated, bringing along the protein-bound DNA fragments, which were thendetected by PCR. The following oligonucleotide pairs were used in the ChIP assays: p21 promoter, 5’-GCTGGCCTGCTGGAACTC-3’/5’-GGCAGCTGCTCACACCTC-3’; Nur77 promoter, 5’-TCACGCGCGCAGACATTCCA-3’/5’-CACCTCTTAAGCGCTCCGTGA-3’; Hsp70 promoter, 5’-CCATGGAGACCAACACCCT-3’/5’-CCCTGGGCTTTTATAAGTCGT-3’.
Gene Expression
RNA was extracted from RNA PAXgene tubes using PAXgene Blood RNA Kit (Qiagen, Valencia, CA). The RNA was reverse transcribed into cDNA using iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA). NanoDrop (Wilmington, DE) spectroscopy was used to quantified cDNA concentration. Dilutions were made with nuclease free water (Ambion, Austin, TX) to bring the final concentration to approximately 35ng/ul.
Gene expression quantification was performed using the iCycler iQ Real-Time PCR detection system (Bio-Rad, Hercules, CA). Product specific primer and probe combinations were used to amplify the genes of interest. A reference gene (YWHAZ) was included in each reaction. Probes were tagged with various fluorophores and Black Hole Quenchers (Biosearch Technologies, Novato, CA). Dark quenchers on a probe prevented fluorescence until the probe anneals with its target sequence. The volume of each qPCR reaction was 25ul, which included 2.0uL of template cDNA, 7.5pmol of each primer and probe (Integrated DNA Technologies, Coralville, IA), and 12.5ul of iQ Multiplex Powermix (Bio-Rad, Hercules, CA). The iQ Multiplex Powermix contains dNTPs, 11mM MgCl2, iTaq DNA polymerase, and stabilizers. The thermal-cycler program consisted of three cycles: 1) 5 minutes at 95°C, 2) 15 seconds at 95°C followed by 30 seconds at 55°C, and 3) 1 minute at 60°C. Cycle 2 was repeated 45 times. The specificity of RT-PCR was controlled by a no-template control.
Gene expression levels were determined using iQ5 software (Bio-Rad, Hercules, CA). Expression was normalized using YWHAZ as a reference gene, and the first time point was used as the baseline.
Validation Summary
Prior to the multiplex reactions, a singleplex SYBR Green reaction with melt curve was used to validate the forward and reverse primers. The product of the SYBR Green reaction was run out on a 2% Agarose (Fisher, Fair Lawn, New Jersey) gel along with pGEM DNA Marker (Promega, Madison, WI) to verify the presence of a single product of the correct size. For multiplex reactions, a six step dilution series (1-32ng/uL) of HepG2 was run in duplicate as a standard curve on each of the plates. The mean coefficient of determination (r2), r2 range, and mean threshold cycle range were calculated from the standard curve for each gene. Correlation coefficients for each gene target ranged from 0.980-0.994.
Statistical Analysis
Gene expression values were summarized with medians and interquartile range (IQR) of the ratios of gene expression relative to baseline. In order to examine whether there was a change in gene expression from baseline, Wilcoxon signed-rank test was used to test the null hypothesis that the median ratio is 1, indicating no change from baseline. For the correlation analysis between Cmax/dose, AUC and gene expression, Spearman's rank correlation coefficient (r) was calculated along with 95% confidence interval. All statistical tests were two-sided, and P < .05 was used to indicate statistical significance. Due to the exploratory nature of this study, no adjustments for multiple comparisons were made. Statistical analysis was performed using SAS® (SAS Institute Inc., Cary, North Carolina) version 9.2 software.
RESULTS
Vorinostat Inhibits Nur77 and p21 Gene Expression in PBMCs
Transcriptional regulation of target genes was evaluated in peripheral blood mononuclear cells (PBMCs) before administration of vorinostat, as well as 2 and 6 hours after administration of vorinostat on days 1 and 9 of cycle 1, when bortezomib was also administered. All 60 subjects had data available for day 1 and 47-49 subjects had data available for day 9. Paired biopsies were evaluated in eleven subjects, although one of the subjects DNA did not amplify and was excluded from analysis.
In PBMCs, Nur77 expression was significantly decreased on both day 1 and day 9 two hours after vorinostat administration relative to baseline, with median ratio of 0.69 (IQR: 0.49-1.04) (p<0.001); 0.28 (0.15-0.7) (p<0.001), respectively, with more a pronounced decrease on day 9. There was a trend for induction of p21 on day 1 two hours after vorinostat administration relative to baseline samples(1.07 (0.85-1.34) (p=0.09)) while it was significantly decreased on day 9 two hours and six hours after administration of vorinostat and bortezomib, 0.67 (0.41-1.03) (p=0.005) vs 0.44 (0.25-1.3) (p=0.007), respectively (See Table 1). HSP70 gene expression was significantly higher after treatment with vorinostat two hours after vorinostat administration on day 1, 1.38 (1.1-1.63) (p<0.001), but not statistically significantly higher on day 9, 1.21 (0.77-1.49). It appears that AKT remained nominally low with statistical significance only on day 9 two hours after administration of vorinostat and bortezomib, 0.83 (0.55-1.17) (p=0.032). Statistically significantinduction of Erbb1 expression was not observed at any time points. Higher gene expression of Erbb2 was seen on day 1 six hours after vorinostat administration, 1.23 (0.78-1.85) (p=0.036); however, this was not shown on day 9.
Table 1.
Gene Expression after Vorinostat Administration, median (1st quartile, 3rd quartile)†
| p21 | HSP70 | Nur 77 | AKT | Erbb1 | Erbb2 | |
|---|---|---|---|---|---|---|
| Biopsies# | ||||||
| Day 9 (n= 10) | 1.13 (0.62, 2.69) | 1.3 (0.72, 1.52) | 0.94 (0.4, 1.49) | 1.16 (0.66, 2.09) | 0.97 (0.35, 1) | 0.89 (0.71, 3.1) |
| Day 1## | ||||||
| 2 hour (n=60) | 1.07 (0.85, 1.34) | 1.38*** (1.1, 1.63) | 0.69*** (0.49, 1.04) | 0.9 (0.7, 1.29) | 0.94 (0.53, 1.54) | 0.88 (0.63, 1.14) |
| 6 hour (n=60) | 1.09 (0.79, 1.33) | 1.04 (0.79, 1.34) | 1.04 (0.79, 1.45) | 0.94 (0.62, 1.25) | 0.86 (0.32, 1.97) | 1.23* (0.78, 1.85) |
| Day 9## | ||||||
| Baseline (n=49) | 0.78** (0.52, 1.22) | 0.93 (0.69, 1.13) | 0.86* (0.41, 1.19) | 0.82* (0.58, 1.18) | 1.09 (0.54, 4.79) | 1.31** (0.97, 2.33) |
| 2 hour (n=48) | 0.67** (0.41, 1.03) | 1.21 (0.77, 1.49) | 0.28*** (0.15, 0.7) | 0.83* (0.55, 1.17) | 1.15 (0.54, 2.66) | 1.06 (0.48, 1.62) |
| 6 hour (n=47) | 0.44** (0.25, 1.3) | 0.98 (0.66, 1.25) | 0.37** (0.21, 1.82) | 0.92 (0.74, 1.4) | 1.13 (0.27, 2.82) | 1 (0.41, 2.5) |
Interquartile range
Gene expression is reported as ratio of gene expression from baseline.
Gene expression is reported as ratio of gene expression from baseline day 1 value.
p< 0.05
p< 0.01
p<0.001
As part of this clinical trial, vorinostat and plasma concentrations of its metabolites were evaluated and have been previously reported (12). Correlations between Cmax/dose and AUC of vorinostat, vorinostat glucuronide and 4-anilino-4-oxobutanoic acid for 48 patients and their gene expression in PBMCs on day 1 of cycle 1 were also evaluated and suggested a trend of consistent positive correlation between plasma dose adjusted Cmax levels of vorinostat and metabolites and p21 expression. Vorinostat (n=48; r=0.27 (−0.02,0.51)); vorinostat glucuronide (n=38; r=0.13 (−0.2, 0.43)); and vorinostat 4-anilino-4-oxobutanoic acid(n=48; r=0.29 (0.01, 0.53)). A similar trend appeared in the correlation between AUC and p21. Vorinostat (n=41; r=0.31 (0, 0.56)); vorinostat glucuronide (n=29; r=0.36 (−0.01, 0.64)); and vorinostat 4-anilino-4-oxobutanoic acid (n=23; r=0.3 (−0.13, 0.63)). There were no significant differences in expression any of the targets between baseline and Day 9 biopsy samples (See Table 1). Correlations between gene expression from PBMC and tissue were examined on day 9 after vorinostat dosing, demonstrating that Nur77 (n=9; r=−0.78 (−0.95, −0.2)) and AKT (n=9; r=−0.81 (−0.95, −0.26)) PBMC expression 6 hours after dosing were inversely correlated with day 9 biopsy expression, p=0.013 and 0.009, respectively. p21, HSP70, Erbb1 and Erbb2 expression was not correlated between tissue and PBMC samples. Analysis of biopsy samples was limited by a small number of samples and variability in gene expression.
Chromatin Precipitation in Paired Tumor Biopsies
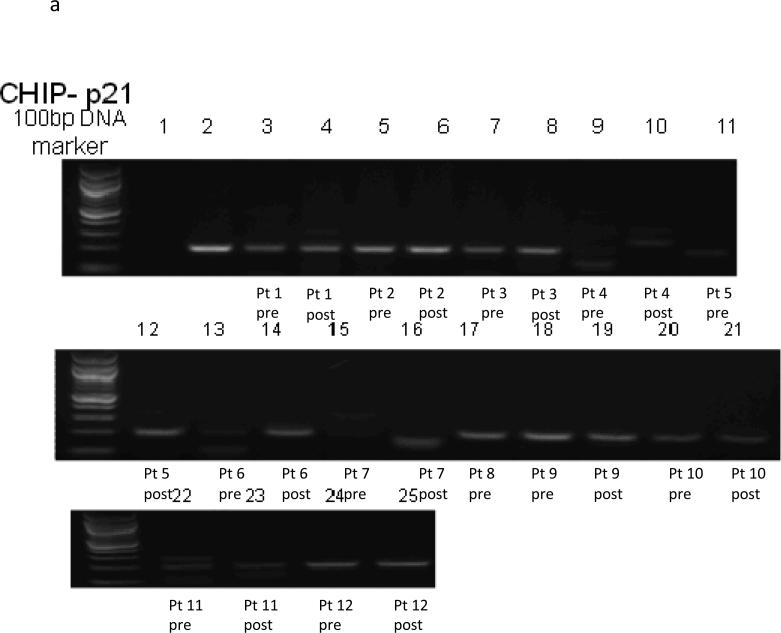
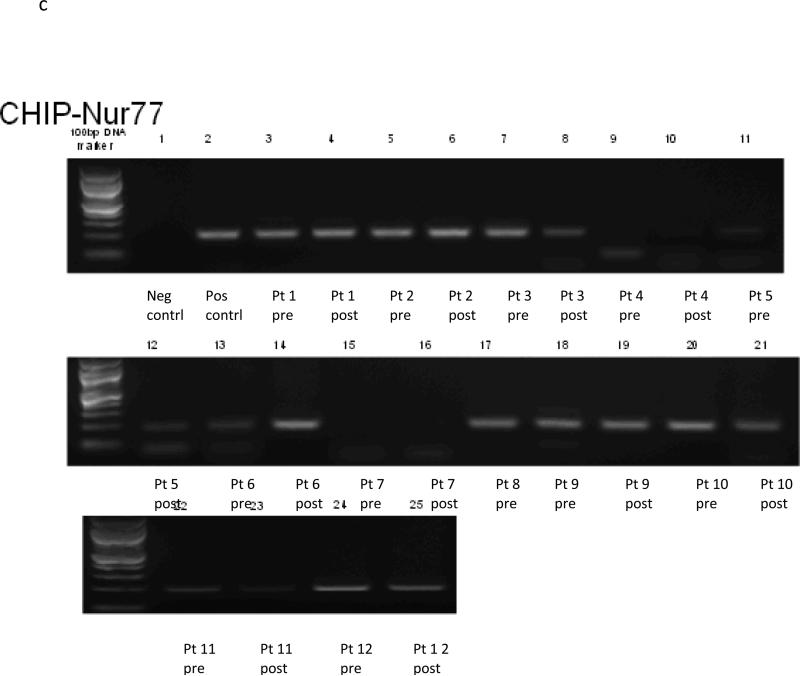
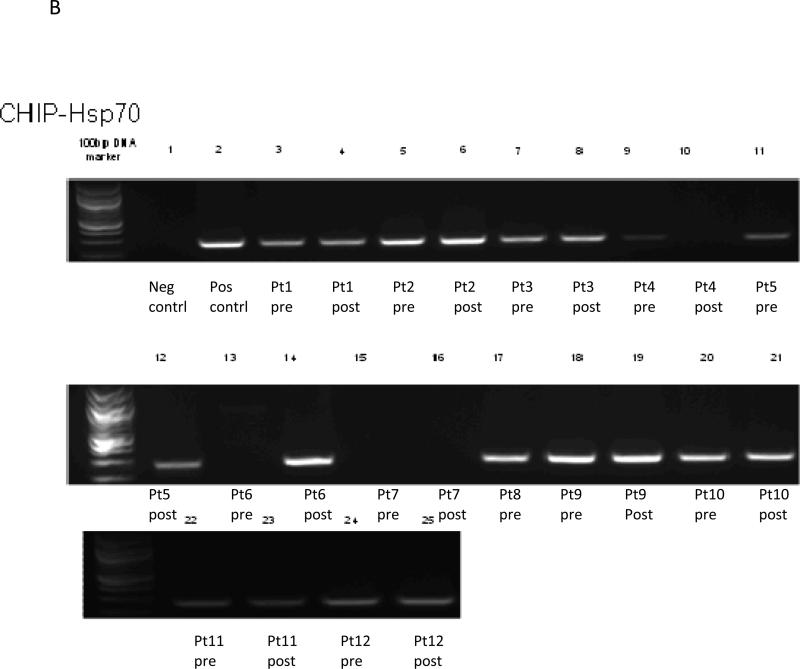
Data for all twelve subjects are included in the ChiP results (See Figure 1a-c). The ChiP assay demonstrates a protein-DNA interaction, in this case interaction of Nur77, HSP70 and p21 with acetylated histone H3, which is transcriptionally active, at baseline and at day 9 after treatment with vorinostat in tissue biopsies.
Figure 1a.
Chip Assay p21
Figure 1c.
Chip Assay of Nur77
Three patients had no transcriptional activation of Nur77 at baseline, with one patient having activation by day 9. Two patients had no activation at either timepoint (patient 4 and 7). Four subjects (Patients 3, 10, 11, 12) had decreases in activation by day 9, with two patients staying the same, and one (patient 6) appearing to have an increase. Effects of vorinostat on Nur77 were inconsistent, with two subjects increasing, four decreasing, two without any expression and three staying about the same.
Four of 11 subjects had no transcriptional activation of p21 at baseline, and two of these (patient 5 and 6) had activation by day 9, showing clear evidence that vorinostat increases transcription of p21 for these two patients. Two patients had no activation at either timepoint (patients 4 and 7). The other seven subjects had transcriptional activation at baseline, with six of these patients either having small increases or stable expression levels, and one (patient 9) appearing to have a small decrease. Overall, 8 of 11 subjects either had increased or consistent transcriptional activation of p21 after vorinostat treatment.
Two patients had no transcriptional activation of HSP70 at baseline, with one patient having activation by day 9. One patient had no activation at either timepoint (patient 7). The other nine subjects had transcriptional activation at baseline, with eight of these patients either having small increases or stable levels, and one (patient 4) appearing to have a small decrease. Similar to p21, 9 of 11 subjects either had increased or consistent transcriptional activation of HSP70 after vorinostat treatment.
DISCUSSION
These results clearly demonstrate in vivo that Nur77 is transcriptionally down regulated by vorinostat, with consistent and statistically significant decreases in gene expression at most time points in PBMCs. p21 was only induced at the earliest timepoint, 2 hours after vorinostat administration, and since its expression is Nur77 dependent (6), this is an expected result. Likewise, AKT, a downstream target of p21, was also downregulated at later timepoints. These findings stand in contrast to prior in vitro evaluations, where treatment with histone deactylase inhibitors, like vorinostat (4,5) and panobinostat (13), resulted in upregulation of Nur77 (13) and p21 (4,5), which are proapoptotic and are one of the postulated mechanisms of action HDAC inhibitors. However, upregulation was observed in transformed cell lines and in cutaneous T cell lymphoma cells. When Chen and colleagues evaluated Nur77 transcription in normal CD34+ cells treated with panobinostat, no upregulation of Nur77 or apoptosis was observed. Thus, our results are consistent with lack of upregulation of Nur77 transcription in normal hematologic cells and may explain the mechanism behind the lack of clinically significant neutropenia observed with HDAC inhibitors, despite having substantial activity in treating cutaneous T cell lymphoma.
HSP70 is a co-chaperone for HSP90 and is required for recruitment of substrates, including AKT, and is known to have anti-apoptotic and cytoprotective functions (14-15). Prior in vitro data have demonstrated induction of HSP70 by vorinostat (16), and in our data, HSP70 is also induced at the earliest time point, 2 hours after vorinostat treatment. HSP90 is a known target of vorinostat, and is inhibited in vitro by vorinostat (17) treatment while HSP70 is known to be induced by HSP90 inhibition (14-15). Therefore, it was initially unclear whether the induction of HSP70 observed in our subjects was a direct effect of vorinostat, or caused by inhibition of HSP90. However, our chromatin immunoprecipitation data suggest that vorinostat interacts directly at the transcriptional level to induce HSP70 gene expression. Given the anti-apoptotic effects of HSP70 induction, we suggest this as a potential mechanism of vorinostat resistance, which should be further evaluated.
All of the quantitative data analyses were performed based on a complete-case analysis with the assumption of non-informative missing data. There were 11-13 subjects whose gene expression data were not available on day 9. As their gene expression levels on day 1 were not significantly different from other subjects available for day 1 to 9, bias due to these missing data is expected to be minimal in this study (data not shown). The primary mechanism of vorinostat is thought to be induction of apoptosis by inducing p21 to halt the cell at the G1 checkpoint (4-6). Gene expression changes were not observed in tissue biopsies, most likely related to tumor heterogeneity and small sample size. Qualitative evidence for transcriptional activation of proapoptotic genes is provided by the chromatin immunoprecipitation, showing that there is an interaction, although variable, between Nur77, p21 and HSP70 and acetylayted H3 in most patients before and after treatment with vorinostat, and that in some cases increases are observed after treatment. Our data support induction of p21 through transcriptional activation as the primary anticancer mechanism of vorinostat at least for some patient .
CONCLUSION
Vorinostat inhibits Nur77 expression, which in turn may decrease p21 and AKT expression in PBMCs. The influence of vorinostat on target gene expression in tumor tissue was variable ;however, most patients demonstrated interaction of acetylated H3 with Nur77, HSP70 and p21 which provides in vivo evidence of interaction with the transcriptionally active acetylated H3 as a mechanism of vorinostat induced apoptosis.
Figure 1B.
Chip Assay of HSP70
ACKNOWLEDGEMENTS
We thank the University of Wisconsin Carbone Cancer Center (UWCCC) Analytical Instrumentation Laboratory for Pharmacokinetics, Pharmacodynamics, & Pharmacogenetics (3P Lab) for acquisition of gene expression and chromatin precipitation data for this research.
We also thank the patients who participated in this clinical trial, and the nurses and research specialist of the UWCCC Phase I Program for their efforts in conducting and managing this trial.
Grant Support: U01 CA062491, Early Clinical Trials of Anti-Cancer Agents with Phase I Emphasis, NCI; CTEP Translational Research Initiative, Contract; and 1UL 1RR025011, Clinical and Translational Science Award, National Center for Research Resources, NIH. The authors would like to thank the University of Wisconsin Carbone Cancer Center (UWCCC) for use of its Shared Services to complete this research. This work is supported in part by NIH/NCI P30 CA014520 to the UW Carbone Cancer Center.
REFERENCES
- 1.Kim HJ, Bae SC. Histone deacetylase inhibitors: molecular mechanisms of action and clinical trials as anti-cancer drugs. Am J Transl Res. 2011 Feb;3(2):166–79. [PMC free article] [PubMed] [Google Scholar]
- 2.Di Marcotullio L, Canettieri G, Infante P, Greco A, Gulino A. Protected from the inside: endogenous histone deacetylase inhibitors and the road to cancer. Biochim Biophys Acta. 2011 Apr;1815(2):241–52. doi: 10.1016/j.bbcan.2011.01.002. Epub 2011 Jan 26. [DOI] [PubMed] [Google Scholar]
- 3.Kavanaugh SM, White LA, Kolesar JM. Vorinostat: A novel therapy for the treatment of cutaneous T-cell lymphoma. Am J Health Syst Pharm. 2010 May 15;67(10):793–7. doi: 10.2146/ajhp090247. [DOI] [PubMed] [Google Scholar]
- 4.Richon VM, Sandhoff TW, Rifkind RA, Marks PA. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc Natl Acad Sci U S A. 2000;97:10014–10019. doi: 10.1073/pnas.180316197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sandor V, Senderowicz A, Mertins S, Sackett D, Sausville E, Blagosklonny MV, Bates SE. P21-dependent g(1)arrest with downregulation of cyclin D1 and upregulation of cyclin E by the histone deacetylase inhibitor FR901228. Br J Cancer. 2000;83:817–825. doi: 10.1054/bjoc.2000.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sarfstein R, Bruchim I, Fishman A, Werner H. The mechanism of action of the histone deacetylase inhibitor vorinostat involves interaction with the insulin-like growth factor signaling pathway. PLoS One. 2011;6(9):e24468. doi: 10.1371/journal.pone.0024468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoon K, Lee SO, Cho SD, Kim K, Khan S, Safe S. Activation of nuclear TR3 (NR4A1) by a diindolylmethane analog induces apoptosis and proapoptotic genes in pancreatic cancer cells and tumor Carcinogenesis. 2011 Jun;32(6):836–42. doi: 10.1093/carcin/bgr040. Epub 2011 Mar 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J, Fiskus W, Eaton K, Fernandez P, Wang Y, Rao R, Lee P, Joshi R, Yang Y, Kolhe R, Balusu R, Chappa P, Natarajan K, Jillella A, Atadja P, Bhalla KN. Cotreatment with BCL-2 antagonist sensitizes cutaneous T-cell lymphoma to lethal action of HDAC7-Nur77-based mechanism. Blood. 2009 Apr 23;113(17):4038–48. doi: 10.1182/blood-2008-08-176024. Epub 2008 Dec 12. [DOI] [PubMed] [Google Scholar]
- 9.Bruzzese F, Leone A, Rocco M, Carbone C, Piro G, Caraglia M, Di Gennaro E, Budillon A. HDAC inhibitor vorinostat enhances the antitumor effect of gefitinib in squamous cell carcinoma of head and neck by modulating ErbB receptor expression and reverting EMT. J Cell Physiol. 2011 Sep;226(9):2378–90. doi: 10.1002/jcp.22574. [DOI] [PubMed] [Google Scholar]
- 10.Lin TY, Fenger J, Murahari S, Bear MD, Kulp SK, Wang D, Chen CS, Kisseberth WC, London CA. AR-42, a novel HDAC inhibitor, exhibits biologic activity against malignant mast cell lines via down-regulation of constitutively activated Kit. Blood. 2010 May 27;115(21):4217–25. doi: 10.1182/blood-2009-07-231985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang QL, Wang L, Zhang YW, Jiang XX, Yang F, Wu WL, Janin A, Chen Z, Shen ZX, Chen SJ, Zhao WL. The proteasome inhibitor bortezomib interacts synergistically with the histone deacetylase inhibitor suberoylanilide hydroxamic acid to induce T-leukemia/lymphoma cells apoptosis. Leukemia. 2009 Aug;23(8):1507–14. doi: 10.1038/leu.2009.41. [DOI] [PubMed] [Google Scholar]
- 12.Schelman WR, Kolesar J, Schell K, Marnocha R, Eickhoff J, Alberti D, Wilding G, Bailey H. A phase I study of vorinostat in combination with bortezomib in refractory solid tumors.. Journal of Clinical Oncology; 2007 ASCO Annual Meeting Proceedings Part I. Vol 25, No. 18S (June 20 Supplement).2007. p. 3573. [Google Scholar]
- 13.Chen J, Fiskus W, Eaton K, et al. Cotreatment with BCL-2 antagonist sensitizes cutaneous T-cell lymphoma to lethal action of HDAC7-Nur77-based mechanism. Blood. 2009 Apr 23;113(17):4038–48. doi: 10.1182/blood-2008-08-176024. Epub 2008 Dec 12. [DOI] [PubMed] [Google Scholar]
- 14.Mossman D, Scott RJ. Long term transcriptional reactivation of epigenetically silenced genes in colorectal cancer cells requires DNA hypomethylation and histone acetylation. PLoS One. 2011;6(8):e23127. doi: 10.1371/journal.pone.0023127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joly AL, Wettstein G, Mignot G, Ghiringhelli F, Garrido C. Dual role of heat shock proteins as regulators of apoptosis and innate immunity. J Innate Immun. 2010;2(3):238–47. doi: 10.1159/000296508. [DOI] [PubMed] [Google Scholar]
- 16.Powers MV, Jones K, Barillari C, Westwood I, van Montfort RL, Workman P. Targeting HSP70: the second potentially druggable heat shock protein and molecular chaperone? Cell Cycle. 2010 Apr 15;9(8):1542–50. doi: 10.4161/cc.9.8.11204. [DOI] [PubMed] [Google Scholar]
- 17.Faraco G, Pancani T, Formentini L, Mascagni P, Fossati G, Leoni F, Moroni F, Chiarugi A. Pharmacological inhibition of histone deacetylases by suberoylanilide hydroxamic acid specifically alters gene expression and reduces ischemic injury in the mouse brain. Mol Pharmacol. 2006 Dec;70(6):1876–84. doi: 10.1124/mol.106.027912. [DOI] [PubMed] [Google Scholar]
- 18.Mühlenberg T, Zhang Y, Wagner AJ, Grabellus F, Bradner J, Taeger G, Lang H, Taguchi T, Schuler M, Fletcher JA, Bauer S. Inhibitors of deacetylases suppress oncogenic KIT signaling, acetylate HSP90, and induce apoptosis in gastrointestinal stromal tumors. Cancer Res. 2009 Sep 1;69(17):6941–50. doi: 10.1158/0008-5472.CAN-08-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marx C, Yau C, Banwait S, Zhou Y, Scott GK, Hann B, Park JW, Benz CC. Proteasome-regulated ERBB2 and estrogen receptor pathways in breast cancer. Mol Pharmacol. 2007 Jun;71(6):1525–34. doi: 10.1124/mol.107.034090. [DOI] [PubMed] [Google Scholar]
- 20.Marx C, Berger C, Xu F, Amend C, Scott GK, Hann B, Park JW, Benz CC. Validated high-throughput screening of drug-like small molecules for inhibitors of ErbB2 transcription. Assay Drug Dev Technol. 2006 Jun;4(3):273–84. doi: 10.1089/adt.2006.4.273. [DOI] [PubMed] [Google Scholar]
- 21.Codony-Servat J, Tapia MA, Bosch M, Oliva C, Domingo-Domenech J, Mellado B, Rolfe M, Ross JS, Gascon P, Rovira A, Albanell J. Differential cellular and molecular effects of bortezomib, a proteasome inhibitor, in human breast cancer cells. Mol Cancer Ther. 2006 Mar;5(3):665–75. doi: 10.1158/1535-7163.MCT-05-0147. [DOI] [PubMed] [Google Scholar]
- 22.Kesarwala AH, Samrakandi MM, Piwnica-Worms D. Proteasome inhibition blocks ligand-induced dynamic processing and internalization of epidermal growth factor receptor via altered receptor ubiquitination and phosphorylation. Cancer Res. 2009 Feb 1;69(3):976–83. doi: 10.1158/0008-5472.CAN-08-2938. Epub 2009 Jan 27. [DOI] [PubMed] [Google Scholar]