Abstract
Among shrimp viral pathogens, white spot syndrome virus (WSSV) and yellow head virus (YHV) are the most lethal agents, causing serious problems for both the whiteleg shrimp, Penaeus (Litopenaeus) vannamei, and the black tiger shrimp, Penaeus (Penaeus) monodon. Another important virus that infects P. vannamei is infectious myonecrosis virus (IMNV), which induces the white discoloration of affected muscle. In the cases of taura syndrome virus and Penaeus stylirostris densovirus (PstDNV; formerly known as infectious hypodermal and hematopoietic necrosis virus), their impacts were greatly diminished after the introduction of tolerant stocks of P. vannamei. Less important viruses are Penaeus monodon densovirus (PmDNV; formerly called hepatopancreatic parvovirus), and Penaeus monodon nucleopolyhedrovirus (PemoNPV; previously called monodon baculovirus). For freshwater prawn, Macrobrachium rosenbergii nodavirus and extra small virus are considered important viral pathogens. Monoclonal antibodies (MAbs) specific to the shrimp viruses described above have been generated and used as an alternative tool in various immunoassays such as enzyme-linked immunosorbent assay, dot blotting, Western blotting and immunohistochemistry. Some of these MAbs were further developed into immunochromatographic strip tests for the detection of WSSV, YHV, IMNV and PemoNPV and into a dual strip test for the simultaneous detection of WSSV/YHV. The strip test has the advantages of speed, as the result can be obtained within 15 min, and simplicity, as laboratory equipment and specialized skills are not required. Therefore, strip tests can be used by shrimp farmers for the pond-side monitoring of viral infection.
Keywords: Shrimp viruses, Immunological assay, Western blot, Immunohistochemistry, Immunochromatographic strip test
Core tip: Monoclonal antibodies (MAbs) specific to various shrimp viruses were generated. The MAbs can be used to detect viral infection in shrimp by immunological assays such as Western blotting, dot blotting, and immunohistochemistry. Some of the MAbs were used to developed immunochromatographic strip tests for specific detection of white spot syndrome virus, yellow head virus, infectious myonecrosis virus, and Penaeus monodon nucleopolyhedrovirus formerly known as monodon baculovirus. The strip test has the advantages of speed, as the result can be obtained within 15 min, and simplicity, as laboratory equipment and specialized skills are not required.
INTRODUCTION
According to the Food and Agriculture Organization of the United Nations FAO report, aquaculture is one of the fastest-growing animal food-producing sectors, and in the next decade, total production from both capture and aquaculture will exceed that of beef, pork or poultry[1]. In 2009, crustaceans contributed approximately 11.2 million tons to global fishery and aquaculture production. During the period of 2000-2008, crustacean production increased at an average annual rate of approximately 15%, faster than in the previous decade. The rapid increase largely reflected the remarkable increase in the whiteleg shrimp, Penaeus vannamei, in China, Thailand and Indonesia[2].
It has been estimated that 40% of tropical shrimp production is lost annually due to infectious diseases[3]. Approximately 60% of the disease-associated loss in shrimp production can be due to viral diseases and 20% to bacterial diseases. The remaining 20% of the loss was attributed to other pathogens, including parasites and fungi[4]. In the case of shrimp viruses, white spot syndrome virus (WSSV) is the most serious pathogen because it is lethal to all cultivated penaeid shrimp species, and mortality can be high and rapid. Another severe pathogen is yellow head virus (YHV), which can cause rapid mortality in P. vannamei and black tiger shrimp (Penaeus monodon). Other important viruses for P. vannamei but not for P. monodon are infectious myonecrosis virus (IMNV) and Taura syndrome virus (TSV). Less important viruses are Penaeus monodon densovirus [PmDNV; formerly called hepatopancreatic parvovirus (HPV)] and Penaeus monodon nucleopolyhedrovirus [PemoNPV; previously called monodon baculovirus (MBV)] because the proper washing of eggs and/or nauplii in the hatchery can eliminate the viruses. Penaeus stylirostris densovirus (PstDNV), formerly known as infectious hypodermal and hematopoietic necrosis virus (IHHNV), can cause high mortality in American blue shrimp (P. stylirostris) and stunt growth in P. vannamei. At present, the commercial stocks of P. vannamei used in Asia are highly tolerant to TSV[4].
For the detection of shrimp viruses, the World Animal Health Organization (the OIE-Office International des Epizooties) recommends polymerase chain reaction (PCR)-based methods[5]. In this brief review, we present alternative assays for the detection of various shrimp viruses based on immunologically developed methods. In all cases, the immunological-based assays are virus specific with optimum sensitivity. Further development into immunochromatographic strip tests that can be used by shrimp farmers to monitor certain shrimp virus infections is discussed.
WSSV
WSSV is a causative agent of white spot disease, which is one of the most devastating diseases in cultured penaeid shrimp, including black tiger shrimp (P. monodon) and whiteleg shrimp (P. vannamei). WSSV infections results in the gross sign of white inclusions embedded in the shrimp cuticle at the late stages of infection[6]. This gross sign of infection was first recognized during an outbreak in Penaeus japonicus in 1993[7]. WSSV is a large, enveloped, rod-shaped, double-stranded DNA virus with a genome size of approximately 300 kbp. The WSSV genome encodes at least 181 open reading frames (ORFs), and most of the predicted gene products show no similarity to known proteins[8]. Due to its unique characteristics, the International Committee on Taxonomy of Viruses (ICTV) classified WSSV as the only member of the genus Whispovirus within a new family called Nimaviridae[9].
At present, at least 40 WSSV structural proteins have been identified, ranging in size from 68 to 6077 amino acid residues[8]. When WSSV virions were subjected to gradient SDS-PAGE analysis, major protein bands were identified and named VP664, VP28, VP26, VP24, VP19 and VP15. VP28 and VP19 were identified as enveloped proteins[10,11], whereas VP26 and VP24 were identified as tegument proteins[8]. Based on solubilization in salt-containing Triton X-100, the tegument protein may loosely associate with both the envelope and nucleocapsid[12]. The nucleocapsid protein VP15 demonstrates DNA-binding activity and may be involved in the packaging of the WSSV genome into the nucleocapsid[11,13]. VP664 is another major nucleocapsid protein with a calculated molecular mass of 664 kDa, and it is encoded by an intron-less ORF of 18234 nucleotides. It is the largest known viral structural protein[14].
Because VP28 is the most abundant WSSV envelope protein[8], it is a good candidate protein for WSSV detection by immunological-based assays. In 2001, four monoclonal antibodies (MAbs) raised against purified WSSV were generated, and they were reacted with the VP28 protein by Western blot analysis. Two of the MAbs were selected for further development into various serological methods, i.e., an immunohistochemical assay and Western blotting for WSSV detection. The two MAbs did not cross-react with hemolymph from shrimp infected with other viruses, including IHHNV, YHV and TSV. However, in that report, no comparative study with PCR was performed[15].
In 2002, three reports described the production of antibodies specific to WSSV VP28 protein. Liu et al[16] produced MAbs using a recombinant 6x-histidine-tagged VP28 as an antigen. The MAbs were used to develop an antigen-capture enzyme-linked immunosorbent assay (ELISA) (Ac-ELISA), and the results revealed that the sensitivity of the Ac-ELISA (400 pg of purified WSSV protein) was comparable to that of PCR (300 pg of DNA extracted from purified WSSV). You et al[17] generated polyclonal antibodies raised against a truncated histidine-tagged VP28 protein, but the antibody cross-reacted with a shrimp protein at 80 kDa. Anil et al[18] reported the production of MAbs against purified WSSV, and the obtained MAbs recognized both the 28 and 18 kDa WSSV proteins. The limit of detection of the immunodot test was 500 pg of the viral protein, which is similar to that of a 1-step PCR assay. However, the immunodot test detected WSSV in only 21 of the 22 PCR-positive samples.
Another study based on the production of MAbs using a truncated VP28 envelope protein lacking the N-terminal transmembrane region as an antigen has also been reported[19]. A MAb named W29 was used to develop immunohistochemical and dot blot assays for WSSV detection, and this MAb could be used to detect WSSV in experimentally infected P. monodon at 12 h. The W29 MAb was further used to develop an immunochromatographic strip test that can be used conveniently by shrimp farmers; the results of this test can be obtained within 15 min without the requirement of sophisticated tools. The strip test employed the W29 MAb conjugated with colloidal gold at a glass fiber located downstream of the sample pad and a rabbit anti-recombinant VP28 antibody combined with the W28 MAb at the test line (Figure 1). However, the sensitivity of the strip test was much lower than that of a 1-step PCR assay[20]. Another immunochromatographic assay called Shrimple® demonstrated lower sensitivity (34.7% of the inoculated shrimp) when compared with real-time PCR (100% of the specimens)[21].
Figure 1.
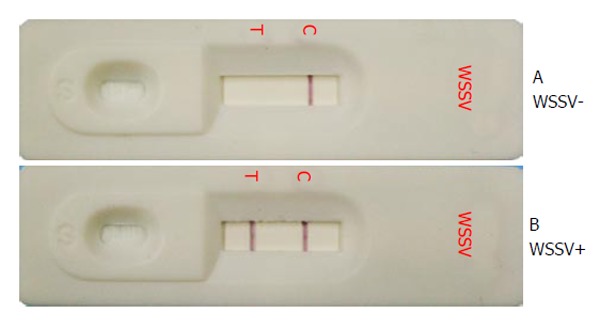
White spot syndrome virus immunochromatographic test strip. Gill homogenates from (A) uninfected P. monodon showing a negative result with only one reddish-purple band at the C line and (B) WSSV-infected P. monodon showing a positive result with two reddish purple bands at the T line and C line. T: Test line; C: Control line. WSSV: White spot syndrome virus.
The second generation of immunochromatographic strip tests, called the dual strip test, was developed for the simultaneous detection of WSSV and YHV[22]. MAbs designated W1 and W30 were raised against a truncated VP28 envelope protein and were used for the development of the dual strip test. The W30 MAb was conjugated to colloidal gold, and the W1 MAb was used at the test line (Figure 2). The detection of WSSV by the dual strip test was approximately 500-fold less sensitive when compared with a 1-step PCR assay. However, the dual strip test could remain functional under storage at room temperature for at least 2 years. At present, this dual strip test has been commercialized by Marine Leader Co. Ltd, Thailand.
Figure 2.
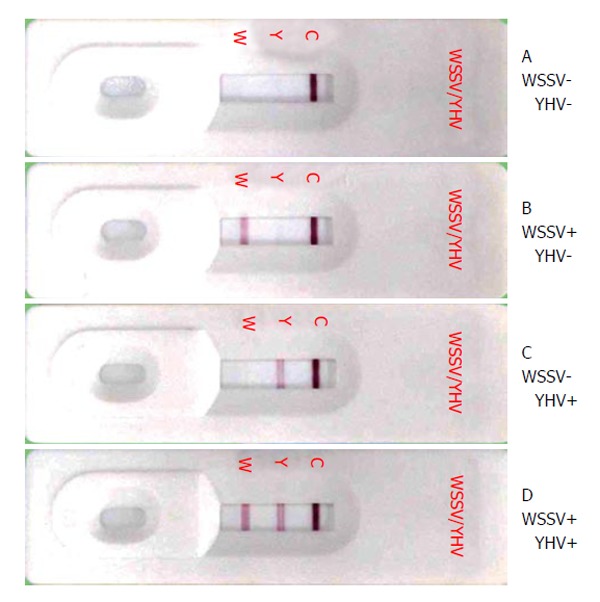
White spot syndrome virus and yellow head virus Dual test strip results. Pleopod homogenates samples from (A) uninfected P. vannamei, (B) WSSV infected P. vannamei, (C) YHV infected P. vannamei or (D) a combination of B and C were applied to the test strip. W: The test line for WSSV, Y: The test line for YHV and C: The control line. WSSV: White spot syndrome virus; YHV: Yellow head virus.
In addition to VP28, other WSSV structural proteins, such as VP19 and VP26, have also been used as targets for WSSV detection. In the case of the VP19 envelope protein, polyclonal and MAbs were raised against a maltose-binding protein (MBP)-VP19 fusion protein[23,24]. The detection limit of MAbs specific to VP19, designated W25, was 1.2 fmole/μL of purified recombinant MBP-VP19 protein, as determined by dot blotting. It has been shown that the combination of MAbs specific to VP19 and VP28 (W29) results in twofold higher sensitivity than if either MAb is used alone. However, the sensitivity of the combined MAbs was still much lower than that of a 1-step PCR assay[24]. In the case of the VP26 structural protein, the polyclonal antibody raised against a truncated 6x-His-tagged VP26 protein demonstrated specific immunoreactivity to viral antigen, as determined by immunohistochemistry and Western blotting[25]. A MAb specific to the VP26 protein is under investigation in our laboratory.
As mentioned above, all of the target proteins for WSSV detection belong to viral structural proteins. However, one report has stated that the ICP11 protein, a non-structural protein, is likely to be a better indicator of WSSV infection. ICP11 is the most highly expressed protein in WSSV-infected gill tissue at 48 h post-infection[26]. ICP11 acts as a DNA mimic by binding to the histone H3 protein, thus disrupting nucleosome assembly[27]. MAbs specific to ICP11 were recently generated using C-terminally intein-tagged ICP11 (ICP11-intein) and N-terminally glutathione-S-transferase (GST)-tagged ICP11 (GST-ICP11) as antigens. The detection limit of the MAbs was approximately 0.7 fmole/μL of GST-ICP11 as determined by dot blotting. A combination of MAbs specific to ICP11, VP28 (W29) and VP19 (W25) increased the detection limit for WSSV to a sensitivity 250-fold lower than that of a 1-step PCR assay[28]. Thus, the development of a higher-sensitivity immunochromatographic strip test for WSSV detection could be achieved in the near future using these MAbs.
TSV
TSV is a major viral pathogen in cultured whiteleg shrimp (P. vannamei). It was first recognized in Ecuador in 1992[29] and later spread to many countries, including those in the Americas, Taiwan, China, Thailand and Indonesia[30-33]. TSV infections can cause gross pathology in P. vannamei that can occur in two phases, the acute and recovery phases. The acute phase can be characterized by a reddish necrotic area on the tail fan, whereas the recovery phase often includes black cuticular lesions in the regions where the acute phase necrosis occurred[6]. TSV is a small, nonenveloped icosahedron, positive-sense, single-stranded RNA virus with a diameter of 32 nm[34]. The viral genome is 10205 nucleotides and contains two large ORFs. ORF1 contains the sequence motifs for helicase, a protease and an RNA-dependent RNA polymerase (RdRp). ORF2 contains the sequences of structural proteins, including three major capsid proteins, VP1 (55 kDa), VP2 (40 kDa) and VP3 (24 kDa), and one minor capsid protein (58 kDa) (Mari 2002). TSV has been classified by the ICTV in the novel genus Aparavirus in a new family Dicistroviridae (in the Order Picornavirales)[35].
VP1 displays greater variation in its amino acid sequence (3.5%) than VP2 and VP3 (both 0.8%)[36]. Therefore, the VP1 region can be used to establish the genetic relationship among TSV isolates. At present, at least four genotypic variants have been identified according to the sequence of the VP1 (equivalent to capsid protein 2; CP2) structural protein. They are the Mexico, Southeast Asia, Belize/Nicaragua and Venezuela/Aruba lineages[37]. Therefore, strain variation may result in an inaccurate diagnosis of TSV infection. Chicken and mouse polyclonal antisera and MAbs against purified TSV antigens have been produced[38]. A MAb specific to VP1 (called 1A1) has been obtained and used to develop Western blot, dot blot and immunohistochemical assays for TSV detection. However, the 1A1 MAb does not recognize TSV isolates from Mexico, Nicaragua and Belize[39-41] and reacts weakly with TSV from Venezuela[42]. Moreover, the 1A1 MAb, raised against purified TSV, displays cross-reactivity to the hemolymph of shrimp infected with PstDNV, YHV or WSSV[38].
In 2006, polyclonal antibodies against VP1 and VP3 were generated using recombinant VP1 and VP3 proteins as the antigens. The obtained MAbs demonstrated specificity to TSV by Western blot and immunohistochemistry[43]. Later, MAbs specific to the VP3 capsid protein were developed and shown to detect TSV infection by dot blot and Western blot without cross-reactions with other shrimp viruses[44]. To increase the sensitivity of TSV detection, MAbs raised against recombinant VP2 proteins were generated[45]. The combination of VP2 and VP3 MAbs can detect TSV infections in field samples of P. vannamei with a better detection limit than a single MAb[45]. Recently, MAbs specific to VP1 of TSV were produced and demonstrated dot blot sensitivity at 2 fmole/μL of GST-VP1[46]. In the near future, an immunochromatographic strip test using MAbs specific to VP1 and VP2 should be used to enhance the pond-side identification of TSV infection.
YHV
YHV is the causative agent of yellow head disease in penaeid shrimp. The virus was named after its gross signs of disease, including a yellowish cephalothorax and a very pale overall coloration of moribund, infected shrimp[6]. YHV first emerged in farmed black tiger shrimp (P. monodon) in Thailand in 1990[47] and caused a loss of shrimp production equivalent to 30-40 million USD[48]. The entire crop is typically lost within a few days after the appearance of the gross signs of this disease[49]. YHV is a bacilliform, enveloped, (+) single-stranded RNA virus classified in the new virus genus Okavirus, the new family Roniviridae and the order Nidovirales[50-52]. There are at least six distinct genetic lineages (genotypes) of YHV, but only YHV type-1 has caused major disease-related losses[53]. Gill-associated virus (GAV or YHV-type 2) has been linked to disease outbreaks with a less severe condition described as mid-crop mortality syndrome[54,55]. Other genotypes were detected exclusively as low level infections in apparently healthy shrimp[53].
Purified YHV virions contain three major structural proteins with molecular masses of 116 kDa (gp116), 64 kDa (gp64) and 20 kDa (p20). The gp116 and gp64 enveloped glycoproteins are encoded by ORF3, whereas the p20 nucleoprotein is encoded by ORF2[56,57]. For YHV detection, a dot blot assay using antiserum against purified YHV was first developed in the year 2000[58]. A MAb named V3-2B that is specific to gp116 was generated and demonstrated specificity to YHV-infected shrimp by dot blot, Western blot and immunohistochemistry[59]. Later, four groups of MAbs specific to gp116, gp64 and p20 were produced against purified YHV virions and were shown to detect YHV infection by dot blot and immunohistochemistry[60]. A single-chain variable fragment antibody directed against gp116 was generated, and a dot blot assay demonstrated its specificity to YHV without cross-reactivity with WSSV and TSV proteins[61].
A convenient immunochromatographic strip test was also developed using a MAb specific to p20 and polyclonal antibodies raised against recombinant p20 protein. The sensitivity of this strip test was approximately 500-fold lower than that of the 1st step reverse transcription (RT)-PCR assay. This kit can also be used to detect GAV infection because the MAb cross-reacts well with GAV[62]. Further improvement of this dual strip test for WSSV and YHV was performed. The sensitivity of the improved dual strip test was 1000-fold lower than an RT-PCR test for YHV and 500-fold lower than a one-step PCR test for WSSV. Although the dual strip test kit has a lower sensitivity than that of PCR, it has advantages in speed and simplicity, as it does not require equipment[22].
IMNV
The first disease outbreak caused by IMNV was reported in farmed Pacific white shrimp (P. vannamei) from Brazil in the year 2004. The gross signs of disease include focal to extensive necrotic areas in skeletal muscle tissues, primarily in the distal abdominal segments and the tail fan, and the appearance of white discoloration of affected muscle[63,64]. Mortality due to IMNV infection in cultivated P. vannamei can reach 70%[65]. IMNV is a nonenveloped, icosahedral virus with a diameter of 40 nm. The genome consists of a double-stranded RNA molecule of 7560 nucleotides containing two ORFs, ORF1 and ORF2. The first half of ORF1 encodes an RNA-binding motif, and the second half encodes a capsid protein with a molecular mass of 160 kDa. ORF2 encodes a putative RdRp[66].
For immunodiagnostic assays, MAbs against a recombinant capsid protein comprising amino acids 300-527 were generated. In an immunodot-blot assay and Western blotting, the MAbs effectively bound tissue extracts from shrimp naturally infected with IMNV. Viral inclusions can also be revealed by immunohistochemistry using these MAbs[67]. Another study on MAb production has also been reported. The gene encoding the capsid protein was amplified into three parts, namely CP-N (nucleotides 2248-3045), CP-I (nucleotides 3046-3954) and CP-C (nucleotides 3955-4953). Two MAbs (IMN7 and IMN12) specific to CP-N and one MAb (IMC1) specific to CP-C were then obtained. The detection sensitivities, as determined by dot blot, were 6 fmole/μL of purified recombinant CP-N protein and 8 fmole/μL of purified recombinant CP-C protein. A combination of all three MAbs resulted in a twofold increase in the detection limit compared to the use of any single MAb. However, the sensitivity is 10-fold lower than that of the a-step RT-PCR assay[68].
Subsequently, the MAbs IMN7 and IMC6, specific to CP-N and CP-C, respectively, were utilized for immunochromatographic strip test production (Figure 3). The sensitivity of the test was comparable to that of dot blot but was approximately 300-fold less sensitive than a one-step RT-PCR[69].
Figure 3.
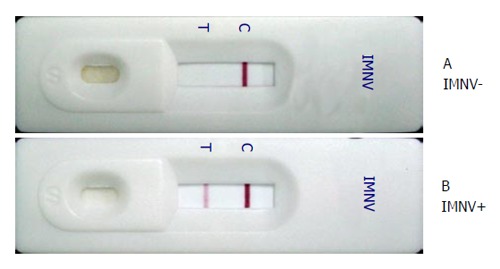
Infectious myonecrosis virus test strip results. Shrimp muscle homogenate samples from (A) an uninfected P. vannamei showing a negative result with only one reddish-purple band at the C line and (B) an IMNV infected P. vannamei showing a positive result with two reddish purple bands. IMNV: Infectious myonecrosis virus.
PemoNPV
PemoNPV was previously called MBV. It has been shown that PemoNPV does not typically cause mortality in farmed P. monodon[70], but it has been linked to stunting, causing the mean length of PemoNPV-infected shrimp to be significantly shorter than that of uninfected shrimp from the same pond[71].
In contrast to insect baculoviruses, little data exist on the genes and genomes of crustacean baculoviruses. However, the PemoNPV genome size has been proposed to fall within the typical baculovirus range of 80 to 160 kb[72]. At present, the only characterized PemoNPV gene is a polyhedrin-encoding gene, and its deduced amino acid sequence revealed no homology to other known proteins[73].
For use in an immunodiagnostic assay, MAbs were raised against a partially purified polyhedrin protein and demonstrated specific PemoNPV detection by Western blot and immunohistochemistry[74]. Subsequently, MAbs raised against a recombinant polyhedrin protein were also produced. Dot blot, Western blot and immunohistochemical assays showed specific PemoNPV detection without cross-reactivity with WSSV, TSV, YHV or PmDNV. Dot blotting using a combination of four MAbs obtained from both studies was approximately 100-fold less sensitive than a 1-step PCR assay[75]. An immunochromatographic strip test using four MAbs raised against partially purified polyhedrin protein was developed and demonstrated a sensitivity 200-fold lower than that of a 1-step PCR assay[76] (Figure 4).
Figure 4.
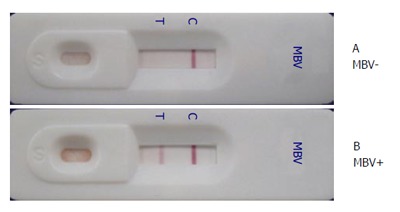
Penaeus monodon nucleopolyhedrovirus immunochromatographic strip test results. Homogenates of (A) uninfected P. monodon postlarvae showing a negative result with only one band at the C line and (B) PemoNPV-infected P. monodon postlarvae displaying a positive result. PemoNPV: Penaeus monodon nucleopolyhedrovirus; MBV: Monodon baculovirus.
PmDNV
PmDNV, also called HPV, is a pathogen responsible for stunted growth in black tiger shrimp (P. monodon). Most PmDNV-infected shrimp grow very slowly and stop growing at approximately 6 cm in length[6,71]. PmDNV is a non-enveloped icosahedral virus that is 22-23 nm in diameter and contains linear ssDNA. It belongs to the family Parvoviridae in the densovirus group[77,78]. Two Asian types of PmDNV have been characterized at the molecular level. One type has been identified in P. chinensis from South Korea[77], P. monodon from Madagascar, Mozambique and Tanzania and P. merguiensis from New Caledonia[79] and Australia[80]. The other type has been identified in infected P. monodon from Thailand and India[78,81]. The complete genome of PmDNV isolated from infected P. monodon in Thailand consists of 6321 nucleotides, representing three ORFs and two non-coding termini. ORF3 encodes a capsid protein of approximately 92 kDa, which may later be cleaved after the first or second arginine residue to produce a 57 kDa or 54 kDa structural protein, respectively[82].
MAbs raised against purified PmDNV isolated from Thailand have been produced, four of which react with a 54 kDa protein by Western blotting and to intranuclear inclusion bodies in tubule epithelial cells in PmDNV-infected tissue by immunohistochemistry[83]. The recombinant capsid proteins of PmDNV have also been used to generate MAbs, and five MAbs have been obtained. The most sensitive MAb displayed a detection limit of 50 fmole/μL of a recombinant protein, as determined by dot blotting. However, the combination of three MAbs revealed a sensitivity 25000-fold lower than a one-step PCR assay[84].
PstDNV
PstDNV, formerly called IHHNV, is a viral pathogen that can cause mortality in juveniles and sub-adults of the blue shrimp P. stylirostris[85] and cuticular deformities and growth retardation (collectively called runt-deformity syndrome) in P. vannamei[86,87]. PstDNV is a non-enveloped icosahedral virus that is 22-23 nm in diameter and contains 4.1 kb of linear ssDNA[85-89]. PstDNV is classified in the family Parvoviridae, subfamily Densovirinae in the genus Brevidensovirus[90]. The PstDNV genome consists of three large ORFs in which the left ORF encodes the non-structural protein, and the right ORF represents the capsid protein; the function of the middle ORF is still unknown[91].
An early report on MAb production described six IgM MAbs generated against purified PstDNV. These MAbs displayed specificity to purified PstDNV preparations in both ELISA and immunoblot assays. However, the ELISA using these MAbs reacted nonspecifically to shrimp samples that were negative for PstDNV by histology and DNA hybridization[92]. In 2009, MAbs raised against a recombinant capsid protein were produced and demonstrated PstDNV specificity in Western blot, dot blot and immunohistochemical assay without cross-reactivity with uninfected shrimp. The sensitivity was 300 pg/μL of recombinant capsid protein, as determined by immunodot-blotting; however, the sensitivity of the dot blot was 1000-fold lower than that of a one-step PCR assay[93].
MrNV NODAVIRUS AND EXTRA SMALL VIRUS
MrNV is an important viral agent that causes white tail disease (WTD) in the giant freshwater prawn Macrobrachium rosenbergii. WTD causes significant mortality in hatchery- and nursery-reared postlarvae[94]. The disease was first reported in Guadeloupe Island (French West Indies) in 1997[95] and was later reported in China[96], India[97], Thailand[98], Taiwan[99] and Australia[100]. MrNV is a non-enveloped, icosahedral virus with a diameter of 26-27 nm and a genome comprised of two pieces of positive-sense ssRNA. RNA 1 is 3202 bp in length and encodes the RdRp, whereas RNA 2 is 1175 bp in length and encodes a viral capsid protein of 43 kDa[96,101]. Extra small virus (XSV) is usually associated with MrNV. XSV is also a non-enveloped, icosahedral virus; it is 15 nm in diameter and contains a 796 bp ssRNA genome[101,102]. XSV has been hypothesized to be a satellite virus that depends on the RdRp of MrNV for replication[96].
For use in immunodiagnostic assays, polyclonal antibodies were raised against a purified viral suspension. A sandwich ELISA was developed and successfully used to identify tissue extracts infected with MrNV as well as purified viral extracts[103]. The purified virus was also used to generate MAbs that demonstrated specificity to the MrNV 42 kDa capsid protein by Western blotting. A triple antibody ELISA (TAS-ELISA) has also been developed and shown to be more sensitive than an indirect ELISA[104]. Recently, a MAb raised against a recombinant MrNV capsid protein was generated and demonstrated a sensitivity of 10 fmole/μL of recombinant protein by dot blotting. However, the sensitivity of this MAb in a dot blot assay using homogenate from naturally MrNV-infected shrimp was 200-fold lower than that of a one-step RT-PCR assay. In the case of XSV, MAbs were raised against a recombinant XSV capsid protein. Four MAbs were obtained and demonstrated a detection limit of 10-20 fmole/μL of purified recombinant protein by dot blotting. Using an XSV-specific MAb and an MrNV-specific MAb, immunohistochemistry on connective tissue sections from prawn with WTD revealed that XSV infection co-localized at varying densities with MrNV infection[105].
ACKNOWLEDGMENTS
We would like to thank Srinakharinwirot University, the Thai National Center for Genetic Engineering and Biotechnology, the Thailand Research Fund, Office of Higher Education Commission, Ministry of Education, Thailand for their funding supports to our research works.
Footnotes
P- Reviewers: Kamal SA, Ren XF S- Editor: Qi Y L- Editor: A E- Editor: Liu SQ
References
- 1.FAO The State of World Fisheries and Aquaculture 2012. Rome, FAO [Google Scholar]
- 2.Bondad-Reantaso MG, Subasinghe RP, Josupeit H, Cai J, Zhou X. The role of crustacean fisheries and aquaculture in global food security: past, present and future. J Invertebr Pathol. 2012;110:158–165. doi: 10.1016/j.jip.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 3.Stentiford GD, Neil DM, Peeler EJ, Shields JD, Small HJ, Flegel TW, Vlak JM, Jones B, Morado F, Moss S, et al. Disease will limit future food supply from the global crustacean fishery and aquaculture sectors. J Invertebr Pathol. 2012;110:141–157. doi: 10.1016/j.jip.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 4.Flegel TW. Historic emergence, impact and current status of shrimp pathogens in Asia. J Invertebr Pathol. 2012;110:166–173. doi: 10.1016/j.jip.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 5.World Organization for Animal Health, Paris, France. Manual of Diagnostics Tests for Aquatic Animal Diseases. OIE (Office International des Epizooties) [Google Scholar]
- 6.Flegel TW. Detection of major penaeid shrimp viruses in Asia, a historical perspective with emphasis on Thailand. Aquaculture. 2006;258:1–33. [Google Scholar]
- 7.Takahashi Y, Itami T, Kondo M, Maeda M, Fujii R, Tomonaga S, Supamattaya K, Boonyaratpalin S. Electron microscopic evidence of bacilliform virus infection in kuruma shrimp (Penaeus japonicus) Fish Pathol. 1994;29:121–125. [Google Scholar]
- 8.Van Etten J. Lesser known large dsDNA viruses. Preface. Curr Top Microbiol Immunol. 2009;328:v–vii. [PubMed] [Google Scholar]
- 9.Valk JM, Flegel BTW, Kou GH, Lightner DV, Lo CF, Loh PC, Walker PW. Nimaviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, et al., editors. VIIIth report of the International Committee on Taxonomy of Viruses. Elsevier: Amsterdam; 2005. p. 187–192. [Google Scholar]
- 10.van Hulten MC, Westenberg M, Goodall SD, Vlak JM. Identification of two major virion protein genes of white spot syndrome virus of shrimp. Virology. 2000;266:227–236. doi: 10.1006/viro.1999.0088. [DOI] [PubMed] [Google Scholar]
- 11.van Hulten MC, Reijns M, Vermeesch AM, Zandbergen F, Vlak JM. Identification of VP19 and VP15 of white spot syndrome virus (WSSV) and glycosylation status of the WSSV major structural proteins. J Gen Virol. 2002;83:257–265. doi: 10.1099/0022-1317-83-1-257. [DOI] [PubMed] [Google Scholar]
- 12.Xie X, Xu L, Yang F. Proteomic analysis of the major envelope and nucleocapsid proteins of white spot syndrome virus. J Virol. 2006;80:10615–10623. doi: 10.1128/JVI.01452-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, Xu X, Hew CL. The structure and function of a gene encoding a basic peptide from prawn white spot syndrome virus. Virus Res. 2001;79:137–144. doi: 10.1016/s0168-1702(01)00340-9. [DOI] [PubMed] [Google Scholar]
- 14.Leu JH, Tsai JM, Wang HC, Wang AH, Wang CH, Kou GH, Lo CF. The unique stacked rings in the nucleocapsid of the white spot syndrome virus virion are formed by the major structural protein VP664, the largest viral structural protein ever found. J Virol. 2005;79:140–149. doi: 10.1128/JVI.79.1.140-149.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poulos BT, Pantoja CR, Bradley-Dunlop D, Aguilar J, Lightner DV. Development and application of monoclonal antibodies for the detection of white spot syndrome virus of penaeid shrimp. Dis Aquat Organ. 2001;47:13–23. doi: 10.3354/dao047013. [DOI] [PubMed] [Google Scholar]
- 16.Liu W, Wang YT, Tian DS, Yin ZC, Kwang J. Detection of white spot syndrome virus (WSSV) of shrimp by means of monoclonal antibodies (MAbs) specific to an envelope protein (28 kDa) Dis Aquat Organ. 2002;49:11–18. doi: 10.3354/dao049011. [DOI] [PubMed] [Google Scholar]
- 17.You Z, Nadala EC, Yang J, van Hulten MC, Loh PC. Production of polyclonal antiserum specific to the 27.5 kDa envelope protein of white spot syndrome virus. Dis Aquat Organ. 2002;51:77–80. doi: 10.3354/dao051077. [DOI] [PubMed] [Google Scholar]
- 18.Anil TM, Shankar KM, Mohan CV. Monoclonal antibodies developed for sensitive detection and comparison of white spot syndrome virus isolates in India. Dis Aquat Organ. 2002;51:67–75. doi: 10.3354/dao051067. [DOI] [PubMed] [Google Scholar]
- 19.Chaivisuthangkura P, Tangkhabuanbutra J, Longyant S, Sithigorngul W, Rukpratanporn , S , Menasveta P, Sithigorngul P. Monoclonal antibodies against a truncated viral envelope protein VP28 can detect white spot syndrome virus WSSV infections in shrimp. Sci Asia. 2004;304:359–363. [Google Scholar]
- 20.Sithigorngul W, Rukpratanporn S, Pecharaburanin N, Longyant S, Chaivisuthangkura P, Sithigorngul P. A simple and rapid immunochromatographic test strip for detection of white spot syndrome virus (WSSV) of shrimp. Dis Aquat Organ. 2006;72:101–106. doi: 10.3354/dao072101. [DOI] [PubMed] [Google Scholar]
- 21.Powell JWB, Burge EJ, Browdy CL, Shepard E. Efficiency and sensitivity determination of Shrimple ®, an immunochromatographic assay for white spot syndrome virus (WSSV), using quantitative realtime PCR. Aquaculture. 2006;257:167. [Google Scholar]
- 22.Sithigorngul P, Rukpratanporn S, Chaivisuthangkura P, Sridulyakul P, Longyant S. Simultaneous and rapid detection of white spot syndrome virus and yellow head virus infection in shrimp with a dual immunochromatographic strip test. J Virol Methods. 2011;173:85–91. doi: 10.1016/j.jviromet.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 23.Chaivisuthangkura P, Phattanapaijitkul P, Thammapalerd N, Rukpratanporn S, Longyant S, Sithigorngul W, Sithigorngul P. Development of a polyclonal antibody specific to VP19 envelope protein of white spot syndrome virus (WSSV) using a recombinant protein preparation. J Virol Methods. 2006;133:180–184. doi: 10.1016/j.jviromet.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 24.Chaivisuthangkura P, Longyant S, Rukpratanporn S, Srisuk C, Sridulyakul P, Sithigorngul P. Enhanced white spot syndrome virus (WSSV) detection sensitivity using monoclonal antibody specific to heterologously expressed VP19 envelope protein. Aquaculture. 2010;299:15–20. [Google Scholar]
- 25.Chaivisuthangkura P, Phattanapaijitkul P, Thammapalerd N, Rukpratanporn S, Longyant S, Sithigorngul W, Sithigorngul P. Production of polyclonal antibodies against recombinant VP26 structural protein of white spot syndrome virus (WSSV) Sci Asia. 2006;32:201–204. doi: 10.1016/j.jviromet.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 26.Wang HC, Wang HC, Kou GH, Lo CF, Huang WP. Identification of icp11, the most highly expressed gene of shrimp white spot syndrome virus (WSSV) Dis Aquat Organ. 2007;74:179–189. doi: 10.3354/dao074179. [DOI] [PubMed] [Google Scholar]
- 27.Wang HC, Wang HC, Ko TP, Lee YM, Leu JH, Ho CH, Huang WP, Lo CF, Wang AH. White spot syndrome virus protein ICP11: A histone-binding DNA mimic that disrupts nucleosome assembly. Proc Natl Acad Sci U S A. 2008;105:20758–20763. doi: 10.1073/pnas.0811233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siriwattanarat R, Longyant S, Chaivisuthangkura P, Wangman P, Vaniksampanna A, Sithigorngul P. Improvement of immunodetection of white spot syndrome virus using a monoclonal antibody specific for heterologously expressed icp11. Arch Virol. 2013;158:967–979. doi: 10.1007/s00705-012-1569-3. [DOI] [PubMed] [Google Scholar]
- 29.Jimenez R. Síndrome de Taura (Resumen) Acuacultura del Ecuador. Rev Espec Cámara Nac Acuacult Guayaquil. 1992;1:1–16. [Google Scholar]
- 30.Lightner DV, Redman RM, Hasson KW, Pantoja CR. Taura syndrome in Penaeus vannamei (Crustacea: Decapoda): gross signs, histopathology and ultrastructure. Dis Aquat Org. 1995;21:53–59. [Google Scholar]
- 31.Tu C, Huang HT, Chuang SH, Hsu JP, Kuo ST, Li NJ, Hsu TL, Li MC, Lin SY. Taura syndrome in Pacific white shrimp Penaeus vannamei cultured in Taiwan. Dis Aquat Org. 1999;38:159–161. [Google Scholar]
- 32.Yu CI, Song YL. Outbreaks of Taura syndrome in Pacific white shrimp Penaeus vannamei cultured in Taiwan. Fish Pathol. 2000;35:21–24. [Google Scholar]
- 33.Lien TW, Hsiung HC, Huang CC, Song YL. Genomic similarity of Taura syndrome virus (TSV) between Taiwan and western hemisphere isolates. Fish Pathol. 2002;37:71–75. [Google Scholar]
- 34.Bonami JR, Hasson KW, Mari J, Poulos BT, Lightner DV. Taura syndrome of marine penaeid shrimp: characterization of the viral agent. J Gen Virol. 1997;78(Pt 2):313–319. doi: 10.1099/0022-1317-78-2-313. [DOI] [PubMed] [Google Scholar]
- 35.Chen YP, Nakashima N, Christian PD, Bakonyi T, Bonning BC, Valles SM, Lightner DV. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier: London; 2012. p. 840–845. [Google Scholar]
- 36.Tang KF, Lightner DV. Phylogenetic analysis of Taura syndrome virus isolates collected between 1993 and 2004 and virulence comparison between two isolates representing different genetic variants. Virus Res. 2005;112:69–76. doi: 10.1016/j.virusres.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 37.Wertheim JO, Tang KF, Navarro SA, Lightner DV. A quick fuse and the emergence of Taura syndrome virus. Virology. 2009;390:324–329. doi: 10.1016/j.virol.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 38.Poulos BT, Kibler R, Bradley-Dunlop D, Mohney LL, Lightner DV. Production and use of antibodies for the detection of Taura syndrome virus in penaeid shrimp. Dis Aquat Org. 1999;37:99–106. [Google Scholar]
- 39.Robles-Sikisaka R, Garcia DK, Klimpel KR, Dhar AK. Nucleotide sequence of 3’-end of the genome of Taura syndrome virus of shrimp suggests that it is related to insect picornaviruses. Arch Virol. 2001;146:941–952. doi: 10.1007/s007050170126. [DOI] [PubMed] [Google Scholar]
- 40.Erickson HS, Zarain-Herzberg M, Lightner DV. Detection of Taura syndrome virus (TSV) strain differences using selected diagnostic methods: diagnostic implications in penaeid shrimp. Dis Aquat Organ. 2002;52:1–10. doi: 10.3354/dao052001. [DOI] [PubMed] [Google Scholar]
- 41.Erickson HS, Poulos BT, Tang KF, Bradley-Dunlop D, Lightner DV. Taura syndrome virus from Belize represents a unique variant. Dis Aquat Organ. 2005;64:91–98. doi: 10.3354/dao064091. [DOI] [PubMed] [Google Scholar]
- 42.Côté I, Navarro S, Tang KFJ, Noble B, Lightner DV. Taura syndrome virus from Venezuela is a new genetic variant. Aquaculture. 2008;284:62–67. [Google Scholar]
- 43.Chaivisuthangkura P, Tejangkura T, Rukpratanporn S, Longyant S, Sithigorngul W, Sithigorngul P. Polyclonal antibodies specific for VP1 and VP3 capsid proteins of Taura syndrome virus (TSV) produced via gene cloning and expression. Dis Aquat Organ. 2006;69:249–253. doi: 10.3354/dao069249. [DOI] [PubMed] [Google Scholar]
- 44.Longyant S, Poyoi P, Chaivisuthangkura P, Tejangkura T, Sithigorngul W, Sithigorngul P, Rukpratanporn S. Specific monoclonal antibodies raised against Taura syndrome virus (TSV) capsid protein VP3 detect TSV in single and dual infections with white spot syndrome virus (WSSV) Dis Aquat Organ. 2008;79:75–81. doi: 10.3354/dao01885. [DOI] [PubMed] [Google Scholar]
- 45.Chaivisuthangkura P, Longyant S, Hajimasalaeh W, Sridulyakul P, Rukpratanporn S, Sithigorngul P. Improved sensitivity of Taura syndrome virus immunodetection with a monoclonal antibody against the recombinant VP2 capsid protein. J Virol Methods. 2010;163:433–439. doi: 10.1016/j.jviromet.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 46.Hajimasalaeh W, Longyant S, Chaivisuthangkura P, Sithigorngul P. Improved immunodetection of Taura syndrome virus using a monoclonal antibody specific for heterologously expressed VP1 capsid protein. Arch Virol. 2013;158:77–85. doi: 10.1007/s00705-012-1460-2. [DOI] [PubMed] [Google Scholar]
- 47.Limuswan C. Handbook for Cultivation of Black Tiger Prawns. Tansetakit: Bangkok; 1991. [Google Scholar]
- 48.Lightner DV, Redman RM. Shrimp diseases and current diagnostic methods. Aquaculture. 1998;164:201–220. [Google Scholar]
- 49.Chantanachookin C, Boonyaratanapalin S, Kasornchandra J, Direkbusarakom S, Ekpanithanpong U, Supamataya K, Siurairatana S, Flegel TW. Histology and ultrastructure reveal a new granulosis-like virus in Penaeus monodon affected by “yellow-head” disease. Dis Aquat Org. 1993;17:145–157. [Google Scholar]
- 50.Cowley JA, Dimmock CM, Spann KM, Walker PJ. Gill-associated virus of Penaeus monodon prawns: an invertebrate virus with ORF1a and ORF1b genes related to arteri- and coronaviruses. J Gen Virol. 2000;81:1473–1484. doi: 10.1099/0022-1317-81-6-1473. [DOI] [PubMed] [Google Scholar]
- 51.Cowley JA, Walker PJ. The complete genome sequence of gill-associated virus of Penaeus monodon prawns indicates a gene organisation unique among nidoviruses. Arch Virol. 2002;147:1977–1987. doi: 10.1007/s00705-002-0847-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sittidilokratna N, Hodgson RA, Cowley JA, Jitrapakdee S, Boonsaeng V, Panyim S, Walker PJ. Complete ORF1b-gene sequence indicates yellow head virus is an invertebrate nidovirus. Dis Aquat Organ. 2002;50:87–93. doi: 10.3354/dao050087. [DOI] [PubMed] [Google Scholar]
- 53.Wijegoonawardane PK, Cowley JA, Phan T, Hodgson RA, Nielsen L, Kiatpathomchai W, Walker PJ. Genetic diversity in the yellow head nidovirus complex. Virology. 2008;380:213–225. doi: 10.1016/j.virol.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spann KM, Cowley JA, Walker PJ, Lester RJG. A yellow-head like virus from Penaeus monodon cultured in Australia. Dis Aquat Org. 1997;31:169–179. [Google Scholar]
- 55.Walker PJ, Cowley JA, Spann KM, Hodgson RAJ, Hall MR, Withychumnarnkul B. Yellow head complex viruses: transmission cycles and topographical distribution in the Asia-Pacific region. In: Browdy CL, Jory DE, et al., editors. The new wave: Proceedings of the Special Session on Sustainable Shrimp Culture, Aquaculture 2001. The World Aquaculture Society: Baton Rouge; 2001. p. 227–237. [Google Scholar]
- 56.Jitrapakdee S, Unajak S, Sittidilokratna N, Hodgson RA, Cowley JA, Walker PJ, Panyim S, Boonsaeng V. Identification and analysis of gp116 and gp64 structural glycoproteins of yellow head nidovirus of Penaeus monodon shrimp. J Gen Virol. 2003;84:863–873. doi: 10.1099/vir.0.18811-0. [DOI] [PubMed] [Google Scholar]
- 57.Sittidilokratna N, Phetchampai N, Boonsaeng V, Walker PJ. Structural and antigenic analysis of the yellow head virus nucleocapsid protein p20. Virus Res. 2006;116:21–29. doi: 10.1016/j.virusres.2005.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nadala EC, Loh PC. Dot-blot nitrocellulose enzyme immunoassays for the detection of white-spot virus and yellow-head virus of penaeid shrimp. J Virol Methods. 2000;84:175–179. doi: 10.1016/s0166-0934(99)00140-8. [DOI] [PubMed] [Google Scholar]
- 59.Sithigorngul P, Chauychuwong P, Sithigorngul W, Longyant S, Chaivisuthangkura P, Menasveta P. Development of a monoclonal antibody specific to yellow head virus (YHV) from Penaeus monodon. Dis Aquat Organ. 2000;42:27–34. doi: 10.3354/dao042027. [DOI] [PubMed] [Google Scholar]
- 60.Sithigorngul P, Rukpratanporn S, Longyant S, Chaivisuthangkura P, Sithigorngul W, Menasveta P. Monoclonal antibodies specific to yellow-head virus (YHV) of Penaeus monodon. Dis Aquat Organ. 2002;49:71–76. doi: 10.3354/dao049071. [DOI] [PubMed] [Google Scholar]
- 61.Intorasoot S, Tanaka H, Shoyama Y, Leelamanit W. Characterization and diagnostic use of a recombinant single-chain antibody specific for the gp116 envelop glycoprotein of Yellow head virus. J Virol Methods. 2007;143:186–193. doi: 10.1016/j.jviromet.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 62.Sithigorngul W, Rukpratanporn S, Sittidilokratna N, Pecharaburanin N, Longyant S, Chaivisuthangkura P, Sithigorngul P. A convenient immunochromatographic test strip for rapid diagnosis of yellow head virus infection in shrimp. J Virol Methods. 2007;140:193–199. doi: 10.1016/j.jviromet.2006.11.034. [DOI] [PubMed] [Google Scholar]
- 63.Lightner DV, Pantoja CR, Poulos BT, Tang KFJ, Redman RM, Andreas T, Bonami JR. Infectious myonecrosis (IMN): a new virus disease of Litopenaeus vannamei. Aquaculture 2004 Book of Abstracts. Baton Rouge, LA: World Aquaculture Society; 2004. p. 353. [Google Scholar]
- 64.Lightner DV, Pantoja CR, Poulos BT, Tang KFJ, Redman RM, Pasos de Andrade T, Bonami JR. Infectious myonecrosis: New Disease in Pacific White Shrimp. Glob Aquac Advocate. 2004;7:85. [Google Scholar]
- 65.Andrade TPD, Redman RM, Lightner DV. Evaluation of the preservation of shrimp samples with Davidson’s AFA fixative for infectious myonecrosis virus (IMNV) in situ hybridization. Aquaculture. 2008;278:179–183. [Google Scholar]
- 66.Poulos BT, Tang KF, Pantoja CR, Bonami JR, Lightner DV. Purification and characterization of infectious myonecrosis virus of penaeid shrimp. J Gen Virol. 2006;87:987–996. doi: 10.1099/vir.0.81127-0. [DOI] [PubMed] [Google Scholar]
- 67.Seibert CH, Borsa M, Rosa RD, Cargnin-Ferreira E, Pereira AM, Grisard EC, Zanetti CR, Pinto AR. Detection of major capsid protein of infectious myonecrosis virus in shrimps using monoclonal antibodies. J Virol Methods. 2010;169:169–175. doi: 10.1016/j.jviromet.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 68.Kunanopparat A, Chaivisuthangkura P, Senapin S, Longyant S, Rukpratanporn S, Flegel TW, Sithigorngul P. Detection of infectious myonecrosis virus using monoclonal antibody specific to N and C fragments of the capsid protein expressed heterologously. J Virol Methods. 2011;171:141–148. doi: 10.1016/j.jviromet.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 69.Chaivisuthangkura P, Senapin S, Wangman P, Longyant S, Sithigorngul P. Simple and rapid detection of infectious myonecrosis virus using an immunochromatographic strip test. Arch Virol. 2013;158:1925–1930. doi: 10.1007/s00705-013-1680-0. [DOI] [PubMed] [Google Scholar]
- 70.Fegan DF, Flegel TW, Sriurairatana S, Waiakruta M. The occurrence, development and histopathology of monodon baculovirus in Penaeus monodon in Southern Thailand. Aquaculture. 1991;96:205–217. [Google Scholar]
- 71.Flegel TW, Nielsen L, Thamavit V, Kongtim S, Pasharawipas T. Presence of multiple viruses in non-diseased, cultivated shrimp at harvest. Aquaculture. 2004;240:55–68. [Google Scholar]
- 72.Mari J, Bonami JR, Poulos B, Lightner DV. Preliminary characterization and partial cloning of the genome of a baculovirus from Penaeus monodon (PmSNPV=MBV) Dis Aquat Org. 1993;163:207–215. [Google Scholar]
- 73.Chaivisuthangkura P, Tawilert C, Tejangkura T, Rukpratanporn S, Longyant S, Sithigorngul W, Sithigorngul P. Molecular isolation and characterization of a novel occlusion body protein gene from Penaeus monodon nucleopolyhedrovirus. Virology. 2008;381:261–267. doi: 10.1016/j.virol.2008.08.036. [DOI] [PubMed] [Google Scholar]
- 74.Boonsanongchokying C, Sang-oum W, Sithigorngul P, Sriurairatana S, Flegel TW. Production of monoclonal antibodies to polyhedrin of monodon baculovirus (MBV) from shrimp. Sci Asia. 2006;32:371–376. [Google Scholar]
- 75.Sridulyakul P, Suwannaka T, Chaivisuthangkura P, Longyant S, Rukpratanporn S, Sithigorngul P. Penaeus monodon nucleopolyhedrovirus detection using monoclonal antibodies specific to recombinant polyhedrin protein. Aquaculture. 2011;321:216–222. [Google Scholar]
- 76.Wangman P, Longyant S, Chaivisuthangkura P, Sridulyakul P, Rukpratanporn S, Sithigorngul P. Penaeus monodon nucleopolyhedrovirus detection using an immunochromatographic strip test. J Virol Methods. 2012;183:210–214. doi: 10.1016/j.jviromet.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 77.Bonami JR, Mari J, Poulos BT, Lightner DV. Characterization of hepatopancreatic parvo-like virus, a second unusual parvovirus pathogenic for penaeid shrimps. J Gen Virol. 1995;76(Pt 4):813–817. doi: 10.1099/0022-1317-76-4-813. [DOI] [PubMed] [Google Scholar]
- 78.Sukhumsirichart W, Wongteerasupaya C, Boonsaeng V, Panyim S, Sriurairatana S, Withyachumnarnkul B, Flegel TW. Characterization and PCR detection of hepatopancreatic parvovirus (HPV) from Penaeus monodon in Thailand. Dis Aquat Organ. 1999;38:1–10. doi: 10.3354/dao038001. [DOI] [PubMed] [Google Scholar]
- 79.Tang KF, Pantoja CR, Lightner DV. Nucleotide sequence of a Madagascar hepatopancreatic parvovirus (HPV) and comparison of genetic variation among geographic isolates. Dis Aquat Organ. 2008;80:105–112. doi: 10.3354/dao01928. [DOI] [PubMed] [Google Scholar]
- 80.La Fauce KA, Elliman J, Owens L. Molecular characterisation of hepatopancreatic parvovirus (PmergDNV) from Australian Penaeus merguiensis. Virology. 2007;362:397–403. doi: 10.1016/j.virol.2006.11.033. [DOI] [PubMed] [Google Scholar]
- 81.Safeena MP, Tyagi A, Rai P, Karunasagar I, Karunasagar I. Complete nucleic acid sequence of Penaeus monodon densovirus (PmDNV) from India. Virus Res. 2010;150:1–11. doi: 10.1016/j.virusres.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 82.Sukhumsirichart W, Attasart P, Boonsaeng V, Panyim S. Complete nucleotide sequence and genomic organization of hepatopancreatic parvovirus (HPV) of Penaeus monodon. Virology. 2006;346:266–277. doi: 10.1016/j.virol.2005.06.052. [DOI] [PubMed] [Google Scholar]
- 83.Rukpratanporn S, Sukhumsirichart W, Chaivisuthangkura P, Longyant S, Sithigorngul W, Menasveta P, Sithigorngul P. Generation of monoclonal antibodies specific to Hepatopancreatic parvovirus (HPV) from Penaeus monodon. Dis Aquat Organ. 2005;65:85–89. doi: 10.3354/dao065085. [DOI] [PubMed] [Google Scholar]
- 84.Srisuk C, Chaivisuthangkura P, Sukhumsirichart W, Sridulyakul P, Longyant S, Rukpratanporn S, Sithigorngul P. Improved immunodetection of Penaeus monodon densovirus with monoclonal antibodies raised against recombinant capsid protein. Aquaculture. 2011;311:19–24. [Google Scholar]
- 85.Lightner DV. Epizootiology, distribution and the impact on international trade of two penaeid shrimp viruses in the Americas. Rev Sci Tech. 1996;15:579–601. doi: 10.20506/rst.15.2.944. [DOI] [PubMed] [Google Scholar]
- 86.Bell TA, Lightner DV. IHHN virus: infectivity and pathogenicity studies in Penaeus stylirostris and Penaeus vannamei. Aquaculture. 1984;38:185–194. [Google Scholar]
- 87.Castille F, Samocha TM, Lawrence AL, He H, Frelier P, Jaenike F. Variability in growth and survival of early postlarval shrimp (Penaeus vannamei Boone 1931) Aquaculture. 1993;113:65–81. [Google Scholar]
- 88.Bonami JR, Adams JR. Unclassified viruses of Crustacea. In: Adams JR, Bonami JR, et al., editors. Atlas of Invertebrate Viruses. CRC Press: Boca Raton, FL; 1991. p. 597–622. [Google Scholar]
- 89.Bonami JR, Trumper B, Mari J, Brehelin M, Lightner DV. Purification and characterization of the infectious hypodermal and haematopoietic necrosis virus of penaeid shrimps. J Gen Virol. 1990;71(Pt 11):2657–2664. doi: 10.1099/0022-1317-71-11-2657. [DOI] [PubMed] [Google Scholar]
- 90.Tijssen P, Agbandje-McKenna M, Almendral JM, Bergoin M, Flegel TW, Hedman K, Kleinschmidt J, Li Y, Pintel DJ, Tattersall P. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier: London; 2012. pp. 405–425. [Google Scholar]
- 91.Shike H, Dhar AK, Burns JC, Shimizu C, Jousset FX, Klimpel KR, Bergoin M. Infectious hypodermal and hematopoietic necrosis virus of shrimp is related to mosquito brevidensoviruses. Virology. 2000;277:167–177. doi: 10.1006/viro.2000.0589. [DOI] [PubMed] [Google Scholar]
- 92.Poulos BT, Lightner DV, Trumper B, Bonami JR. Monoclonal antibodies to a penaeid shrimp Parvovirus, infectious hypodermal and hematopoietic necrosis virus (IHHNV) J Aquat Anim Health. 1994;6:149–154. [Google Scholar]
- 93.Sithigorngul P, Hajimasalaeh W, Longyant S, Sridulyakul P, Rukpratanporn S, Chaivisuthangkura P. Simple immunoblot and immunohistochemical detection of Penaeus stylirostris densovirus using monoclonal antibodies to viral capsid protein expressed heterologously. J Virol Methods. 2009;162:126–132. doi: 10.1016/j.jviromet.2009.07.026. [DOI] [PubMed] [Google Scholar]
- 94.Pillai D, Bonami JR. A review on the diseases of freshwater prawns with special focus on white tail disease of Macrobrachium rosenbergii. Aquaculture Res. 2012;43:1029–1037. [Google Scholar]
- 95.Arcier JM, Herman F, Lightner DV, Redman RM, Mari J, Bonami JR. A viral disease associated with mortalities in hatchery-reared postlarvae of the giant freshwater prawn Macrobrachium rosenbergii. Dis Aquat Org. 1999;38:177–181. [Google Scholar]
- 96.Qian D, Shi Z, Zhang S, Cao Z, Liu W, Li L, Xie Y, Cambournac I, Bonami JR. Extra small virus-like particles (XSV) and nodavirus associated with whitish muscle disease in the giant freshwater prawn, Macrobrachium rosenbergii. J Fish Dis. 2003;26:521–527. doi: 10.1046/j.1365-2761.2003.00486.x. [DOI] [PubMed] [Google Scholar]
- 97.Sahul Hameed AS, Yoganandhan K, Sri Widada JS, Bonami JR. Studies on the occurrence of Macrobrachium rosenbergii nodavirus and extra small virus-like particles associated with white tail disease of M. rosenbergii in India by RT-PCR detection. Aquaculture. 2004;238:127–133. [Google Scholar]
- 98.Yoganandhan K, Sri Widada J, Bonami JR, Sahul Hameed AS. Simultaneous detection of Macrobrachium rosenbergii nodavirus and extra small virus by a single tube, one-step multiplex RT-PCR assay. J Fish Dis. 2005;28:65–69. doi: 10.1111/j.1365-2761.2004.00606.x. [DOI] [PubMed] [Google Scholar]
- 99.Wang CS, Chang JS, Wen CM, Shih HH, Chen SN. Macrobrachium rosenbergii nodavirus infection in M. rosenbergii (de Man) with white tail disease cultured in Taiwan. J Fish Dis. 2008;31:415–422. doi: 10.1111/j.1365-2761.2008.00921.x. [DOI] [PubMed] [Google Scholar]
- 100.Owens L, La Fauce K, Juntunen K, Hayakijkosol O, Zeng C. Macrobrachium rosenbergii nodavirus disease (white tail disease) in Australia. Dis Aquat Organ. 2009;85:175–180. doi: 10.3354/dao02086. [DOI] [PubMed] [Google Scholar]
- 101.Bonami JR, Shi Z, Qian D, Sri Widada J. White tail disease of the giant freshwater prawn, Macrobrachium rosenbergii: separation of the associated virions and characterization of MrNV as a new type of nodavirus. J Fish Dis. 2005;28:23–31. doi: 10.1111/j.1365-2761.2004.00595.x. [DOI] [PubMed] [Google Scholar]
- 102.Widada JS, Bonami JR. Characteristics of the monocistronic genome of extra small virus, a virus-like particle associated with Macrobrachium rosenbergii nodavirus: possible candidate for a new species of satellite virus. J Gen Virol. 2004;85:643–646. doi: 10.1099/vir.0.79777-0. [DOI] [PubMed] [Google Scholar]
- 103.Romestand B, Bonami JR. A sandwich enzyme linked immunosorbent assay (S-ELISA) for detection of MrNV in the giant freshwater prawn, Macrobrachium rosenbergii (de Man) J Fish Dis. 2003;26:71–75. doi: 10.1046/j.1365-2761.2003.00432.x. [DOI] [PubMed] [Google Scholar]
- 104.Qian D, Liu W, Jianxiang W, Yu L. Preparation of monoclonal antibody against Macrobrachium rosenbergii Nodavirus and application of TAS-ELISA for virus diagnosis in post-larvae hatcheries in east China during 2000-2004. Aquaculture. 2006;261:1144–1150. [Google Scholar]
- 105.Longyant S, Senapin S, Sanont S, Wangman P, Chaivisuthangkura P, Rukpratanporn S, Sithigorngul P. Monoclonal antibodies against extra small virus show that it co-localizes with Macrobrachium rosenbergii nodavirus. Dis Aquat Organ. 2012;99:197–205. doi: 10.3354/dao02482. [DOI] [PubMed] [Google Scholar]


