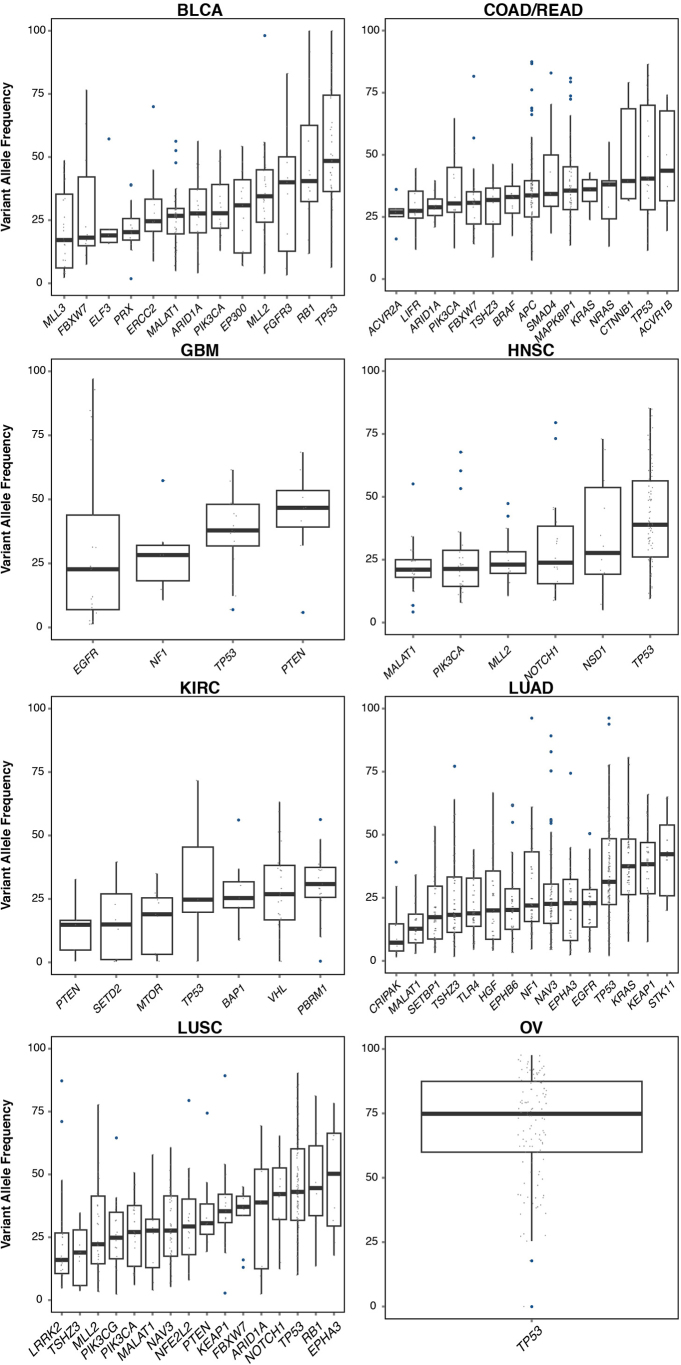
Extended Data Figure 7. VAF distribution of mutations in SMGs across tumours from BLCA, KIRC, HNSC, LUAD, LUSC, COAD/READ, OV and GBM.
To minimize the effect of copy number alterations on VAFs, only mutations residing in copy number neutral segments were used for this analysis. Only mutation sites with ≥20× coverage were used for analysis and plotting. SMGs with at least five data points were included in the plot.