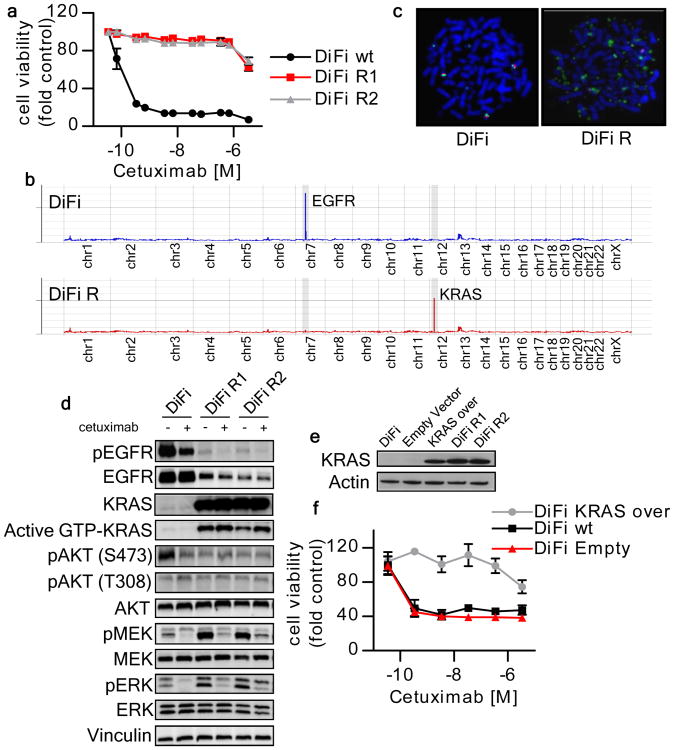
Figure 1. KRAS amplification mediates acquired resistance to cetuximab in DiFi cells.
(a) Parental and cetuximab resistant DiFi cells were treated for one week with increasing concentrations of cetuximab. Cell viability was assayed by the ATP assay. Data points represent means ± SD of three independent experiments. (b) Whole exome gene copy number analysis of parental and cetuximab resistant DiFi cells. Individual chromosomes are indicated on the x axis. The lines indicate the sequencing depth (y axis) over exome windows of 100,000 bp. (c) FISH analysis confirming KRAS amplification in DiFi-R but not parental DiFi cells. KRAS locus BAC DNA (probe RP11-707G18; green) and chromosome 12 paint (red) were hybridized to the metaphase spreads of DiFi cells. (d) DiFi cells were treated with cetuximab 35 nM for 24 hours, after which whole-cell extracts were subjected to Western blot analysis and compared to untreated cells. DiFi R1 and R2 were plated in the absence of cetuximab for 7 days or maintained in their normal growth medium (with cetuximab 35 nM) before protein analysis. Active KRAS (GTP-KRAS) was assessed by GST-Raf1 pull-down. Whole-cell extracts were blotted with phosphor-EGFR (Tyr 1068), total EGFR, total KRAS, phosphor-AKT (Thr 308), phosphor-AKT (Ser473), total AKT, total MEK1/2 and phospho-MEK1/2, total ERK1/2 and phospho-ERK1/2 antibodies. Vinculin was included as a loading control. (e) Western blot analysis of KRAS protein in DiFi cells infected with a KRAS lentivirus. Actin is shown as a loading control (f) Ectopic expression of wild-type KRAS in parental DiFi cells confers resistance to cetuximab.