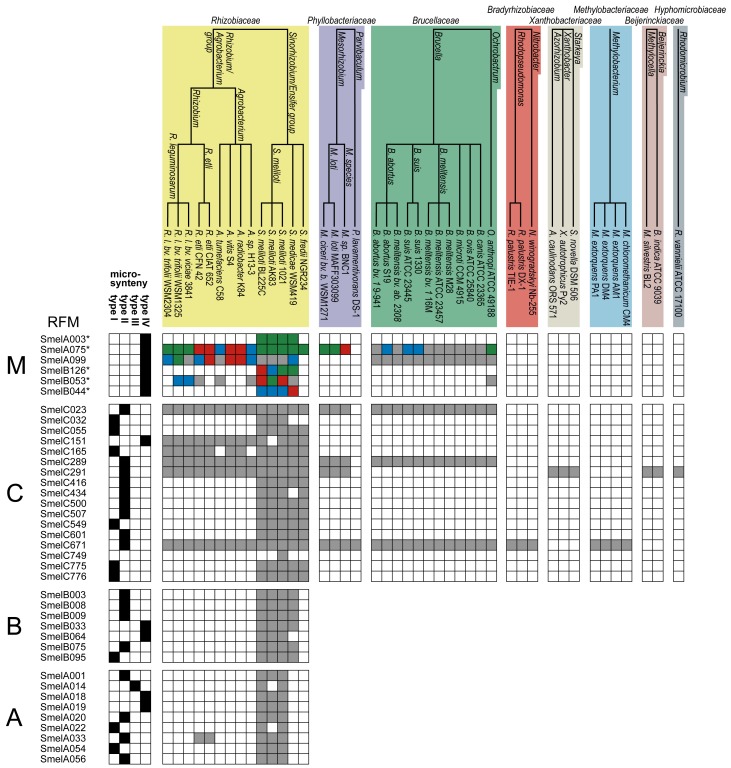
Figure 3.
Distribution pattern of trans-encoded sRNAs in the Rhizobiales. The simplified phylogenetic tree includes sequenced strains and was adopted from the Pathosystems Resource Integration center (PATRIC) [48]. Analyzed bacterial strains that reveal no relatives in each RFM were removed from this scheme. A complete summary of all genomes used in this study is given in Table S2. sRNA occurrence of particular RFMs is given in each line. Chromosome (C), pSymA (A), and pSymB (B) of S. meliloti 1021 carry the initial set of trans-encoded sRNAs used in this comparative study and were indicated as different blocks separated by black, horizontal lines. The upper block (M) summarizes RFMs of sRNAs with several gene copies in particular genomes. * indicates RFMs that contain several sRNA gene copies in the S. meliloti 1021 strain (Table S1). The color code indicates the number of related sRNAs in each strain: 1 = grey, 2 = blue, 3 = red, and ≥4 = green. Complete (type I), extensive (type II), partial (type III), and fragmented (type IV) microsynteny is represented by black boxes.