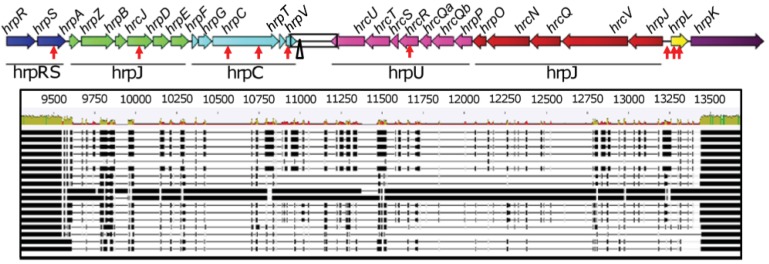
Figure 1.
Organization of Type III Secretion System (TTSS) cluster of Psv5, Psn23 and Psf134. Red arrows represent the single nucleotide polymorphisms (SNPs) found among pathovar TTSS sequences. Black triangle indicates the localization of a partial insertion sequence, remnant of an IS66 element. Names of operons are not in italics to differentiate from gene names. For the part of the sequence highlighted by the black square, corresponding to the hypervariable region between hrpC and hrpU operons, the alignment among the sequences derived from the seventeen Pseudomonas species examined in this study is schematically reported. These species are ordered from top to bottom as: (1) P. savastanoi pv. savastanoi ITM317 (Psv5); (2) P. savastanoi pv. nerii ESC23 (Psn23); (3) P. savastanoi pv. fraxinii NCPPB1006 (Psf134); (4) P. syringae pv. aesculi str. 2250; (5) P. syringae pv. phaseolicola 1448A; (6) P. syringae pv. tabaci ATCC11528; (7) P. syringae pv. syringae str. 61; (8) P. syringae Cit 7; (9) P. syringae pv. syringae B728a; (10) P. syringae pv. aceris M302273PT; (11) P. syringae pv. pisi 1704B; (12) P. syringae pv. aptata DSM50252; (13) P. syringae pv. japonica M301072PT; (14) P. syringae pv. tomato DC3000; (15) P. syringae pv. tagetis LMG5090; (16) P. viridiflava PNA3.3a; (17) P. viridiflava LP23.