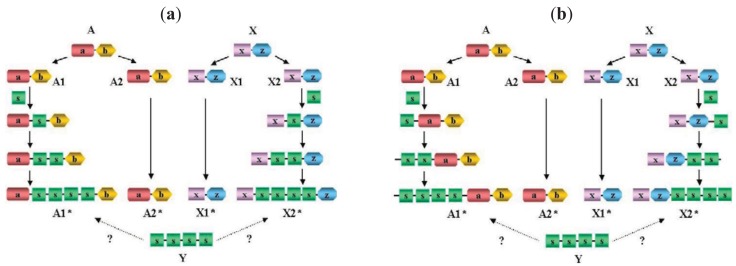
Figure 3.
Domain Architecture evolution: consequences of confusing epaktology and paralogy. Proteins A and X are unrelated ancestral multidomain proteins. During evolution domain shuffling inserts the same domain-type (domain s) into orthologs of A and X proteins independently followed by tandem duplication of this domain, resulting in proteins A1* and X2* in an extant species (Species*). Overall sequence similarity score of proteins A1* and X2* may be so significant that A1* appears to be much more closely related to X2* than its true paralog (A2*), therefore, based on sequence similarity searches it may be concluded that X2* is the closest paralog of A1* (inferring. that A1* and X2* diverged from a common hypothetical ancestor Y) and that independent terminal gain of domains a,b and x,z occurred in the two lines leading to paralogs A1* and X2*. In contrast with this interpretation, A1* is a paralog of A2* (and X1* is a paralog of X2*) and independent gain and duplication of an s domain occurred in both lineages. Note the that although the two scenarios depicted in (a) and (b) differ in the actual events (internal vs. terminal insertion of domain s), if the epaktologs are treated as paralogs the conclusion will be similar in as much as the DA of A1* and X2* differ in terminal positions.