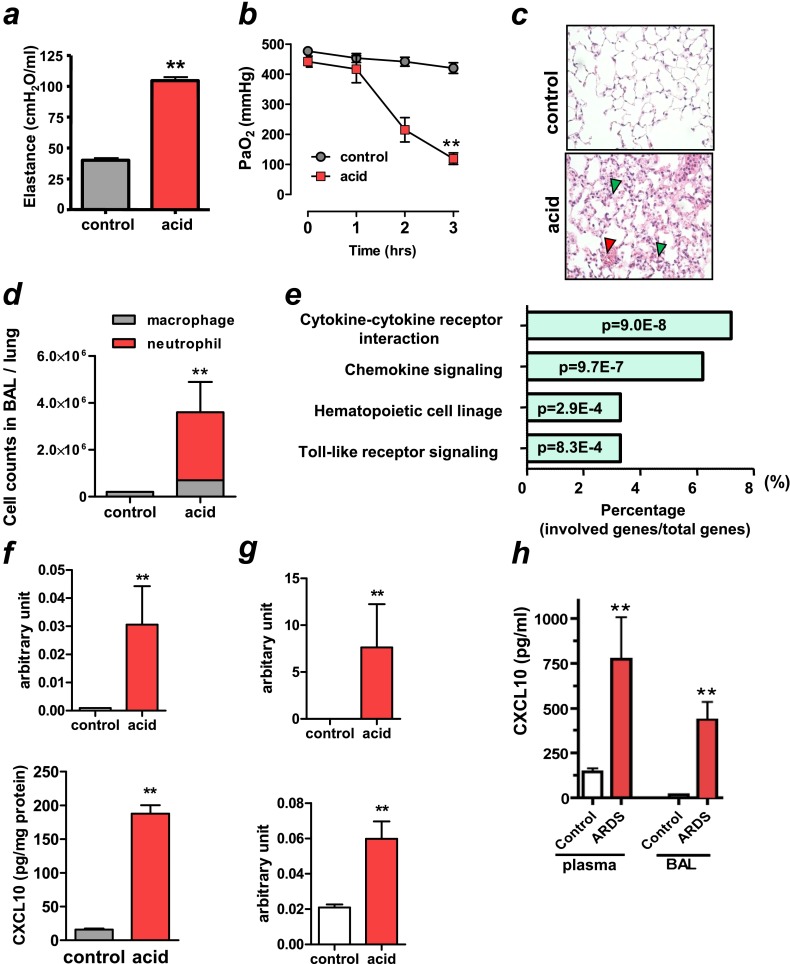
Figure 1.
Elevated CXCL10 expression in nonviral lung injury/acute respiratory distress syndrome (ARDS) in animals and patients. (a–e) After intratracheal instillation of saline (control) or hydrochloric acid (acid) to rats, animals were ventilated for 3 hours. (a) Lung elastance in control and acid treatment at 3 hours. n = 6 each group. **P < 0.01 compared with control rats. (b) Changes in PaO2 after saline or acid aspiration. n = 6 each group. **P < 0.01 for the whole time course comparing to controls. (c) Lung histopathology in control and acid-treated animals at 3 hours. Note inflammatory cell infiltration (green arrowheads) and pulmonary hemorrhage (red arrowhead) in acid-treated lungs. Hematoxylin and eosin staining. Original magnifications ×200. (d) Cell counts in lung lavage fluid in control and acid treatment at 3 hours. n = 4 for control group; n = 6 for acid-treated group. Neutrophil numbers were higher in acid-treated lungs than in control lungs. **P < 0.01 compared with control lungs. (e) KEGG pathway enrichment analysis of the up-regulated genes after acid aspiration. The numbers inside the columns represent the P value of the analysis. (f, g) After intratracheal instillation of saline (control) or hydrochloric acid (acid) to mice, animals were ventilated for 3 hours. (f) CXCL10 mRNA (upper panel) and protein concentration (lower panel) in lungs of control and acid-treated animals at 3 hours. n = 4 for control group; n = 6 for acid-treated group. **P < 0.01 compared with control lungs. (g) CXCL2 (upper panel) and IFN-β1 (lower panel) mRNA expressions in lungs of control and acid-treated animals at 3 hours. n = 4 for control group; n = 6 for acid-treated group. **P < 0.01 compared with control lungs. (h) CXCL10 levels in plasma (left bars) and bronchoalveolar lavage fluid (BAL) (right bars) in control subjects (n = 17) and patients with ARDS with nonviral origins (n = 10). **P < 0.01 compared with control subjects. Data in a, b, d, f–h are presented as mean ± SEM.